Abstract
To examine the expressed gene profile during encystation of Acanthamoeba castellanii Castellani, we used differentially expressed gene (DGE) screening by RT-PCR with 20 sets of random primers. From this analysis, we found that approximately 16 genes showed upregulation during encystation. We chose 6 genes, which had relatively higher expression levels, for further investigation. Based on homology search in database, DEG2 showed 55% of similarity with xylose isomerase, DEG9 showed 37% of similarity with Na P-type ATPase, and DEG14 showed 77% of similarity with subtilisin-like serine proteinase. DEG3 and DEG26 were identified as hypothetical proteins and DEG25 exhibited no significant similarity to any known protein. Encystation of Acanthamoeba has been suggested to be a process to resist adverse environmental or nutritional conditions. Further characterization studies of these genes may provide us with more information on the encystation mechanism of Acanthamoeba.
-
Key words: Acanthamoeba, differentially expressed gene, encystation
Acanthamoeba is an opportunistic pathogen responsible for several diseases in humans, such as granulomatous amebic encephalitis (GAE), amebic keratitis and dermatitis (
Marciano-Cabral and Carbral, 2003).
Acanthamoeba comprises 2 distinct stages, i.e., trophozoite and cyst. The trophozoite is the vegetative form, which conducts movement, phagocytosis, metabolism, and cell division (
Baumann and Murphy, 1995;
Wang and Ahearn, 1997). The cyst is formed from a trophozoite, when desiccation, starvation, or other adverse condition prevails, and is conversed to the trophozoite, when conditions are favorable (
Weisman, 1976;
Cordingley et al., 1996). Because trophozoites can transform to cysts under adverse conditions, such as host immune responses or chemical treatments in human infections (
McClellan et al., 2002), it is difficult to treat the ameba infection with almost all kinds of antibiotics. If it is possible to understand the molecular mechanisms of encystation and to disrupt its process, it will be easier to treat the infection. It will also be applicable to drug development against other cyst forming pathogenic protozoa. To compare the gene expression between trophozoites and cysts, we conducted differentially expressed gene screening with
Acanthamoeba castellanii Castellani (American Type Culture Collections; ATCC #30011, Beltsville, Maryland, USA).
Acanthamoeba was obtained from ATCC and was cultured axenically in PYG medium and induced encystation by the procedure of Bowers and Korn (
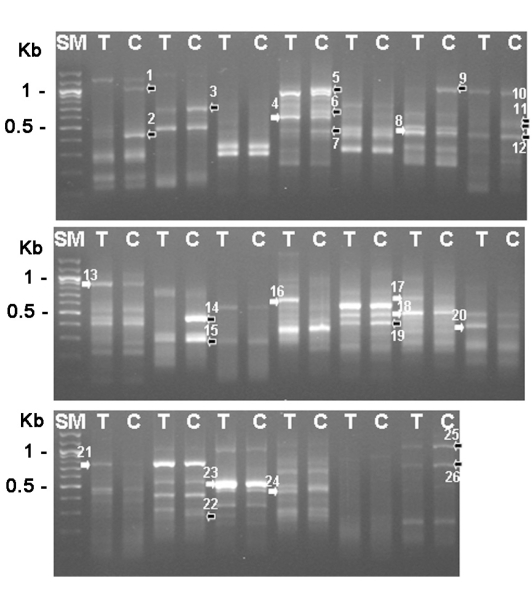
1969). The total RNAs were purified from trophozoites and 3-day cysts, and their cDNAs were made, which was followed by the differentially expressed gene (DEG) screening with 20 sets of random primers (Seegen, Seoul, Korea). From this analysis between trophozoites and cysts, we found that approximately 16 genes were up-regulated and 10 genes were down-regulated during encystation (
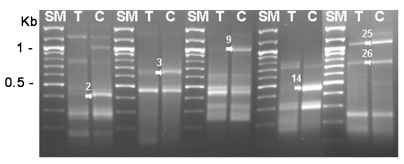
Fig. 1). Then, 6 genes (DEG2, 3, 9, 14, 25 and 26) showing significant up-regulation were confirmed the differential expression between cysts and trophozoites by DEG analysis again (
Fig. 2). The expression pattern of these genes may suggest roles of the gene products in the encystation of
Acanthamoeba. The genes were sequenced followed by submission for blast search (
Table 1). Based on blast search results, the DEG2 protein showed 55% sequence similarity with xylose isomerase, DEG9 showed 37% similarity with Na P-type ATPase, DEG14 showed 77% similarity with subtilisin-like serine proteinase. DEG3 and DEG26 were identified as hypothetical proteins and DEG25 exhibited no significant similarity to any known protein at database.
Interestingly, DEG14 showed significant sequence similarity with subtilisin-like serine proteinase (AhSub), which was reported to be a secretory enzyme and a potential virulence factor (
Hong et al.,2000). Until now, many proteinases of
Acanthamoeba, such as serine, cysteine, and metallo-proteinases, have been suggested to be involved in the pathogenesis or phagocytosis (
Mitro et al., 1994;
Hong et al., 2000;
Kong et al., 2000;
Hong et al., 2002;
Kim et al., 2006;
Serrano-Luna et al., 2006). We identified full length ORF of DEG14 of
A. castellanii by RACE-PCR. Based on the sequence analysis, DEG14 belonged to the serine protease family subtilisin group (data not shown). The deduced amino acid sequence of DEG14 showed 76% similarity with that of AhSub (
Fig. 3). Here, we identified 6 genes induced by encystations, while its expression in the trophozoite is very low or absent. Further studies on these genes may provide more understanding on the encystation mechanisms of
Acanthamoeba.
Notes
-
This work was supported by the Korea Research Foundation Grant funded by Korea Government (MOEHRD, Basic Research Promotion Fund) (KRF-2005-204-E00027) and the Brain Korea 21 Project in 2007.
References
Fig. 1Gel electrophoresis analysis with differentially expressed gene screening results (26 genes). SM; 100 bp ladder, T; trophozoite, C; 3-day cyst, black arrows; reduced genes during encystation, white arrows; increased genes during encystation.

Fig. 2Re-amplification of DEG2, 3, 9, 14, 25 and 26 between cysts and trophozoites. SM; 100 bp ladder, T; trophozoite, C; 3-day cyst, arrows; increased genes during encystation.

Fig. 3Comparision of the deduced amino acid sequences of DEG14 with AhSub (subtilisin-like serine proteinase).

Table 1.Significant matches of DEGs with database sequences of other organisms
Table 1.
|
DEG No. |
Length |
Putative identification |
Organism |
Accession No. |
Database |
Score |
P-value |
|
DEG2 |
314 |
Xylose isomerase |
Reinekea sp. |
ZP 01113810.1 |
ref |
150 |
0.0 × e–9
|
|
DEG3 |
596 |
Hypothetical protein |
Nitrosospira multiformis
|
YP_412886.1 |
ref |
157 |
2.0 × e–9
|
|
DEG9 |
701 |
Putative Na P-type ATPase |
Physcomitrella patens
|
CAD91920.1 |
emb |
283 |
9.0 × e–24
|
|
DEG14 |
383 |
Subtilisin-like serine proteinase |
Acanthamoeba healyi
|
AF221523_1 |
gb |
375 |
7.0 × e–35
|
|
DEG25 |
917 |
No significant similarity found |
|
|
|
|
|
|
DEG26 |
623 |
Hypothetical protein |
Rhodobacter sphaeroides
|
YP_001167477.1 |
ref |
100 |
1.1 × e–2
|
Citations
Citations to this article as recorded by

- Encystment and Excystment Processes in Acanthamoeba castellanii: An Emphasis on Cellulose Involvement
Mathew Choaji, Ascel Samba-Louaka, Zineb Fechtali-Moute, Willy Aucher, Sébastien Pomel
Pathogens.2025; 14(3): 268. CrossRef - Oxidase enzyme genes are differentially expressed during Acanthamoeba castellanii encystment
Christian Q. Scheckhuber, Rebeca Damián Ferrara, Jesús Gómez-Montalvo, Sutherland K. Maciver, Alvaro de Obeso Fernández del Valle
Parasitology Research.2024;[Epub] CrossRef - Ouabain, ATPase inhibitor, potentially enhances the effect of polyhexamethylene biguanide on Acanthamoeba castellanii
Kuang-Yi Shih, Yao-Tsung Chang, Yu-Jen Wang, Jian-Ming Huang
International Journal for Parasitology: Drugs and Drug Resistance.2024; 25: 100550. CrossRef -
Acanthamoeba
keratitis: new hopes for potential interventions for a curable but often refractory disease
Bader Saleem Alawfi, Naveed Ahmed Khan, David Lloyd, Ruqaiyyah Siddiqui
Expert Review of Ophthalmology.2024; 19(4): 271. CrossRef - Biological characteristics and pathogenicity of Acanthamoeba
Yuehua Wang, Linzhe Jiang, Yitong Zhao, Xiaohong Ju, Le Wang, Liang Jin, Ryan D. Fine, Mingguang Li
Frontiers in Microbiology.2023;[Epub] CrossRef - mRNA Sequencing Reveals Upregulation of Glutathione S-Transferase Genes during Acanthamoeba Encystation
Alvaro de Obeso Fernández del Valle, Christian Quintus Scheckhuber, David Armando Chavaro-Pérez, Erandi Ortega-Barragán, Sutherland K. Maciver
Microorganisms.2023; 11(4): 992. CrossRef - Stimulation of Acanthamoeba castellanii excystment by enzyme treatment and consequences on trophozoite growth
Zineb Fechtali-Moute, Philippe M. Loiseau, Sébastien Pomel
Frontiers in Cell and Developmental Biology.2022;[Epub] CrossRef - Peganum harmala Extract Has Antiamoebic Activity to Acanthamoeba triangularis Trophozoites and Changes Expression of Autophagy-Related Genes
Rachasak Boonhok, Suthinee Sangkanu, Julalak Chuprom, Mayuna Srisuphanunt, Roghayeh Norouzi, Abolghasem Siyadatpanah, Farzaneh Mirzaei, Watcharapong Mitsuwan, Sueptrakool Wisessombat, Maria de Lourdes Pereira, Mohammed Rahmatullah, Polrat Wilairatana, Chr
Pathogens.2021; 10(7): 842. CrossRef - Whole Organism Model to Study Molecular Mechanisms of Differentiation and Dedifferentiation
Areeba Anwar, Ruqaiyyah Siddiqui, Naveed Ahmed Khan
Biology.2020; 9(4): 79. CrossRef - New insights into the mechanical properties of Acanthamoeba castellanii cysts as revealed by phonon microscopy
Fernando Pérez-Cota, Richard J. Smith, Hany M. Elsheikha, Matt Clark
Biomedical Optics Express.2019; 10(5): 2399. CrossRef - Encystation: the most prevalent and underinvestigated differentiation pathway of eukaryotes
Pauline Schaap, Christina Schilde
Microbiology.2018; 164(5): 727. CrossRef - Acanthamoeba and mimivirus interactions: the role of amoebal encystment and the expansion of the ‘Cheshire Cat’ theory
Ludmila Karen dos Santos Silva, Paulo Victor Miranda Boratto, Bernard La Scola, Cláudio Antônio Bonjardim, Jônatas Santos Abrahão
Current Opinion in Microbiology.2016; 31: 9. CrossRef - Autophagy protein 12 plays an essential role in Acanthamoeba encystation
So-Hee Kim, Eun-Kyung Moon, Yeonchul Hong, Dong-Il Chung, Hyun-Hee Kong
Experimental Parasitology.2015; 159: 46. CrossRef - Down-Regulation of Cellulose Synthase Inhibits the Formation of Endocysts in Acanthamoeba
Eun-Kyung Moon, Yeonchul Hong, Dong-Il Chung, Youn-Kyoung Goo, Hyun-Hee Kong
The Korean Journal of Parasitology.2014; 52(2): 131. CrossRef - Chloroquine Has a Cytotoxic Effect on Acanthamoeba Encystation through Modulation of Autophagy
Bijay Kumar Jha, Hui-Jung Jung, Incheol Seo, Hyun Ah Kim, Seong-Il Suh, Min-Ho Suh, Won-Ki Baek
Antimicrobial Agents and Chemotherapy.2014; 58(10): 6235. CrossRef - Silencing of xylose isomerase and cellulose synthase by siRNA inhibits encystation in Acanthamoeba castellanii
Yousuf Aqeel, Ruqaiyyah Siddiqui, Naveed Ahmed Khan
Parasitology Research.2013; 112(3): 1221. CrossRef - Acanthamoebadifferentiation: a two-faced drama ofDr Jekyll and Mr Hyde
RUQAIYYAH SIDDIQUI, RICKY DUDLEY, NAVEED AHMED KHAN
Parasitology.2012; 139(7): 826. CrossRef - Protein kinase C signaling molecules regulate encystation of Acanthamoeba
Eun-Kyung Moon, Dong-Il Chung, Yeonchul Hong, Hyun-Hee Kong
Experimental Parasitology.2012; 132(4): 524. CrossRef - Cellular, Biochemical, and Molecular Changes during Encystment of Free-Living Amoebae
Emilie Fouque, Marie-Cécile Trouilhé, Vincent Thomas, Philippe Hartemann, Marie-Hélène Rodier, Yann Héchard
Eukaryotic Cell.2012; 11(4): 382. CrossRef - Microarray Analysis of Differentially Expressed Genes between Cysts and Trophozoites ofAcanthamoeba castellanii
Eun-Kyung Moon, Ying-Hua Xuan, Dong-Il Chung, Yeonchul Hong, Hyun-Hee Kong
The Korean Journal of Parasitology.2011; 49(4): 341. CrossRef - Major Role for Cysteine Proteases during the Early Phase of Acanthamoeba castellanii Encystment
David Leitsch, Martina Köhsler, Martina Marchetti-Deschmann, Andrea Deutsch, Günter Allmaier, Michael Duchêne, Julia Walochnik
Eukaryotic Cell.2010; 9(4): 611. CrossRef - Acanthamoeba castellanii: Proteins involved in actin dynamics, glycolysis, and proteolysis are regulated during encystation
Sabrina Bouyer, Marie-Hélène Rodier, Alain Guillot, Yann Héchard
Experimental Parasitology.2009; 123(1): 90. CrossRef -
Characterization of a Serine Proteinase Mediating Encystation of
Acanthamoeba
Eun-Kyung Moon, Dong-Il Chung, Yeon-Chul Hong, Hyun-Hee Kong
Eukaryotic Cell.2008; 7(9): 1513. CrossRef