Abstract
This report describes the molecular characterization of the Tc8.2 gene of Trypanosoma cruzi. Both the Tc8.2 gene and its encoded protein were analyzed by bioinformatics, while Northern blot and RT-PCR were used for the transcripts. Besides, immunolocalization of recombinant protein was done by immunofluorescence and electron microscopy. Analysis indicated the presence of a single copy of Tc8.2 in the T. cruzi genome and 2-different sized transcripts in epimastigotes/amastigotes and trypomastigotes. Immunoblotting showed 70 and 80 kDa polypeptides in epimastigotes and trypomastigotes, respectively, and a differential pattern of immunolocalization. Overall, the results suggest that Tc8.2 is differentially expressed during the T. cruzi life cycle.
-
Key words: Trypanosoma cruzi, Tc8.2 expression, dual localization
Trypanosoma cruzi is the etiologic agent of Chagas’ disease, also known as American trypanosomiasis. The disease is found mostly in the Americas, and it is estimated that there are at least 10 million infected people and 10-14 thousand deaths each year [
1]. This protozoan has 4 different forms, the epimastigote and metacyclic trypomastigote, located in the insect digestive tract, and the amastigote and bloodstream trypomastigote occurring in the mammalian host. The genome-sequencing project of
T. cruzi was partially completed in 2005 [
2]; however, like other TriTryp genomes, the main problem is the failure to predict the function of most genes that are annotated as “hypothetical proteins”. Moreover, these proteins can also be found as putative, in which the predictions are based on similarity to previously characterized proteins or their functional domains.
Tc8.2 gene was isolated from λZap
T. cruzi trypomastigote cDNA library using 350 bp fragment of β- chemokine CCL2 receptor as a probe (LM Yamauchi, unpublished data). The
Tc8.2 clone was sequenced, and the nucleotide sequence analysis showed no homology to chemokine receptor; however, it revealed similarity to a putative transmembrane transport protein. In view of this, the aim of the study was the characterization of the
Tc8.2 gene and the analysis of the transcript and protein expression.
We used the 1,431 bp nucleotide sequence derived from
Tc8.2 cDNA clone (accession no. AF496671) as query in blastn analysis against the TritrypDB database (
http://tritrypdb.org/tritrypdb). All sequences with alignment >715 bp and similarity >60% were included in this analysis. Northern blot analysis was performed using total RNA (12 µg), isolated from different developmental stages of
T. cruzi; epimastigotes, trypomastigotes (obtained from supernatant of LLCMK
2 cells, 120 hr post-infection), and amastigotes (obtained from 72 hr infected cells). The β-tubulin gene was used as the constitutive control. The epimastigote and trypomastigote RNAs were also used in reverse transcription PCR (RT-PCR). The cDNAs were used for amplification with the oligonucleotides 5ʹatgtctttgaatgcgaacg-3ʹ and 5ʹctaacgactggactttcgcc-3ʹ, specific for the
Tc8.2 gene. To ensure that there was only gene amplification from cDNA molecules and not from genomic DNA, RT-PCR reactions were carried out without the reverse transcriptase enzyme.
Epimastigote DNA of
T. cruzi Y strain (maintained in LIT medium at 28˚C) was extracted [
3] and amplified, on the basis of the consensus sequence of the
Tc8.2 gene, a 1,779-bp fragment by PCR using the following oligonucleotides: 5ʹ-gggacaa gtttgtacaaaaaagcaggctggaaggagataatgtctttgaatgcgaacg-3ʹ and 5ʹ-ggggaccactttgtacaagaaagctgggtcctaacgactggactttcgcc-3ʹ. In addition to the sequence corresponding to the gene, both primers contain recombination sites for cloning into the pDONR vector (Gateway, Invitrogen, Carlsbad, California, USA), which are indicated in bold. The
Tc8.2 gene was recombined into the expression vector pDEST™17 for production of His6-tagged recombinant protein in the
Escherichia coli BL21 (DE) strain. The recombinant protein was used to produce polyclonal antiserum in BALB/c mice, which were subcutaneously immunized 4 times with 10 µg Tc8.2 protein in Freund's adjuvant (Sigma-Aldrich, St. Louis, Missouri, USA). The use of mice was in accordance with the Ethics Committee on Animal Experiments of Faculdade de Medicina de Ribeirão Preto/USP, no. 13/2006.
Anti-Tc8.2 antiserum was used for Western blot [
4] analysis using lysates of 5×10
6 parasites and also for immunolocalization assays. In immunofluorescence assays, parasites (1×10
7) were deposited on glass slides, incubated with anti-Tc8.2 (1:100) in PBS and with AlexaFluor-488-conjugated antimouse IgG (Invitrogen) (1:400). DNA was stained with DAPI (4'6- diamino-2-phenylindole, Invitrogen) (1:1,000) in PBS. Slides were mounted with
n-propyl-gallate (Sigma-Aldrich) and the images acquired with an Olympus BX51 fluorescence microscope. In the ultrastructural analysis, parasites were embedded in Unicryl, and resin polymerization was carried out in BEEM capsules at -20˚C under UV light. Ultrathin sections were obtained with a Leica Ultramicrotome (Wien, Austria) and grids. This was followed by incubation with anti-Tc8.2 serum diluted 1:100 in blocking solution (3% BSA in PBS, pH 8.0). The grids were then incubated with gold-labeled goat anti-mouse IgG (Sigma-Aldrich) diluted 1:200 in blocking solution. Grids were stained with uranyl acetate and lead citrate for further observation in a Jeol JEM 1400 transmission electron microscope (Tokyo, Japan).
To further characterize the Tc8.2 protein, analyses in CBS Prediction Servers and PROSITE were performed to evaluate post-translational modifications such as the presence of transmembrane helix and hydrophobic motifs, as well as phosphorylation and glycosylation sites.
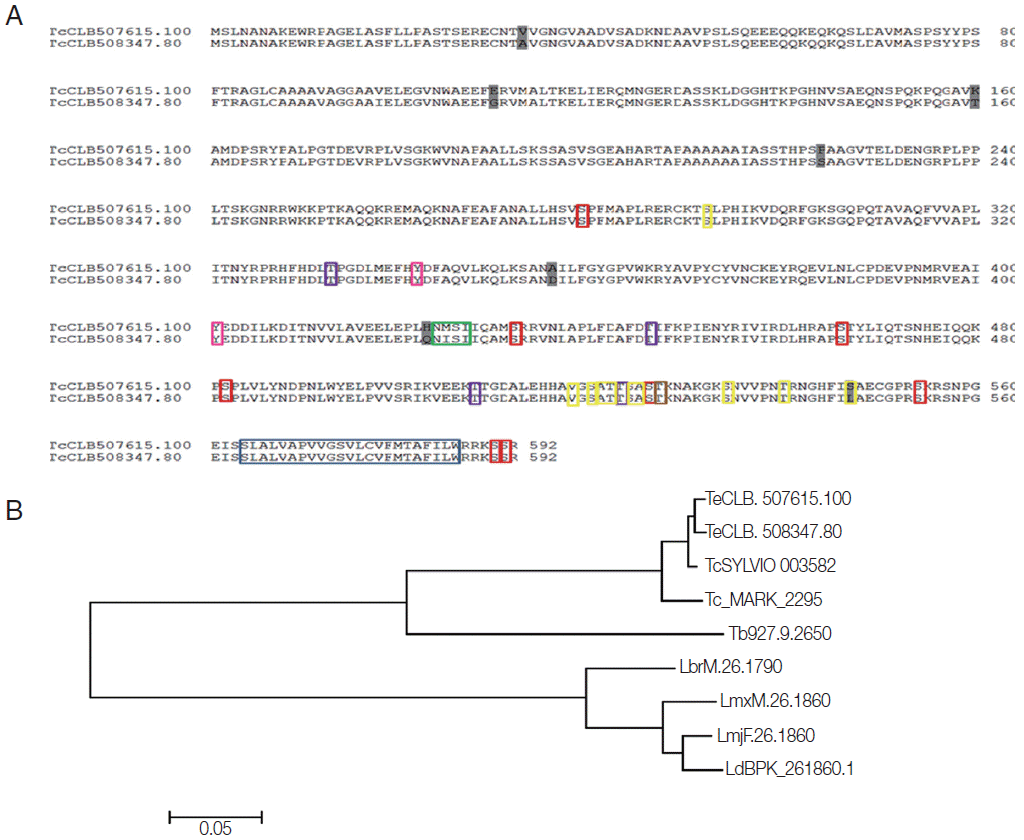
Using bioinformatics analysis, we identified 1 copy of the
Tc8.2 gene in the
T. cruzi Sylvio and Dm28c strains and
T. cruzi marinkellei genome and 2 copies (TcChr 33-P and TcChr 33-S) in the clone CL Brener. Since CL Brener is a hybrid strain, both copies corresponded to full-length sequences of 1,779 bp; moreover, molecular karyotypes also showed 2 chromosomal bands, XIX and X [
5], in this strain. The analysis of the amino acid sequence of these genes (592 aa) in CL Brener showed a high similarity level, with the presence of few nonconserved residues (
Fig. 1A), and analysis with other trypanosomatids suggested a high level of conservation in these species, for example, with
T. cruzi Sylvio and
T. cruzi Marinkellei, which displayed about 90% similarity. In
T. brucei and
Leishmania species (
L. major,
L. brasiliensis,
L. mexicana, and
L. donovani) analyzed, single copies of the
Tc8.2 gene were found, displaying about 60% similarity (
Fig. 1B).
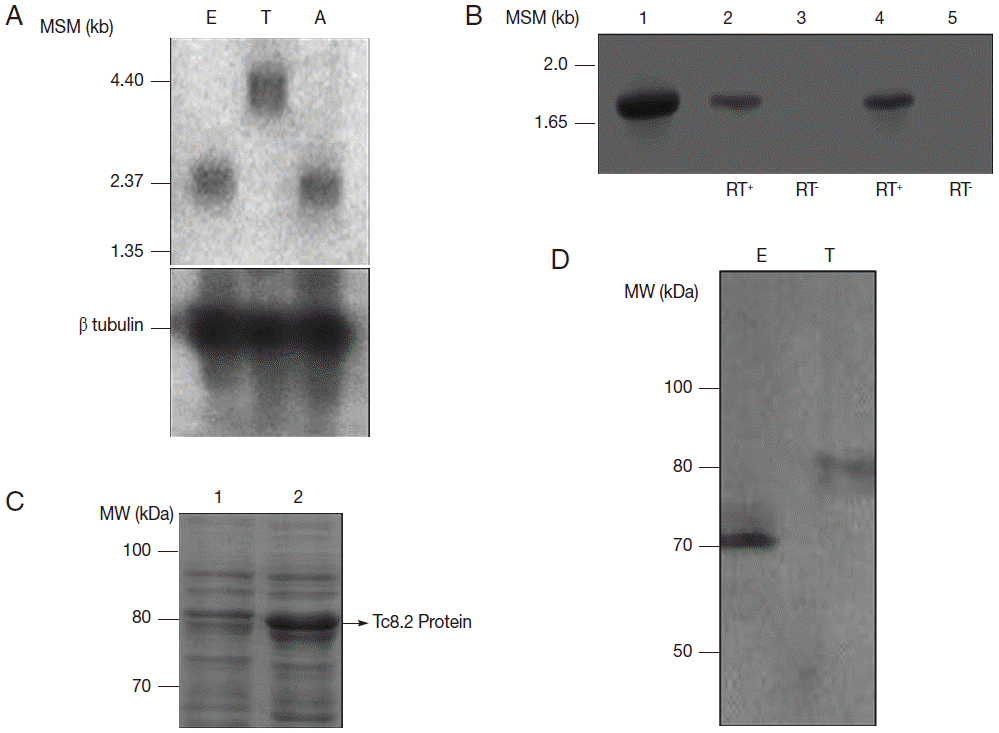
In Northern blot analysis,
Tc8.2 recognized a 3.0 kb transcript in epimastigotes and amastigotes, and 4.4 kb transcript in trypomastigotes. Hybridization with the tubulin gene, a constitutive control, showed the same amount of RNA in the 3 developmental forms (
Fig. 2A). Therefore, we raised the hypothesis that 2 different
Tc8.2 transcripts were produced to generate proteins with different molecular weights in epimastigotes and trypomastigotes. Thus, RT-PCR of epimastigote and trypomastigote RNAs were performed, and comparison was made with amplification of the
Tc8.2 gene using
T. cruzi genomic DNA. The results showed the same-sized amplicon (
Fig. 2B). No amplification was observed in the absence of reverse transcriptase, suggesting the same gene generated 2 different-sized transcripts.
To determine whether this gene encodes proteins with different characteristics, we expressed the
Tc8.2 gene (
Fig. 2C) and immunized mice, and the antiserum recognized a single protein in the epimastigote and trypomastigote extract. Interestingly, the antibodies recognized 2 different-sized proteins; a 70 kDa protein in the epimastigote extract, and a higher molecular weight protein of 80 kDa in the trypomastigote (
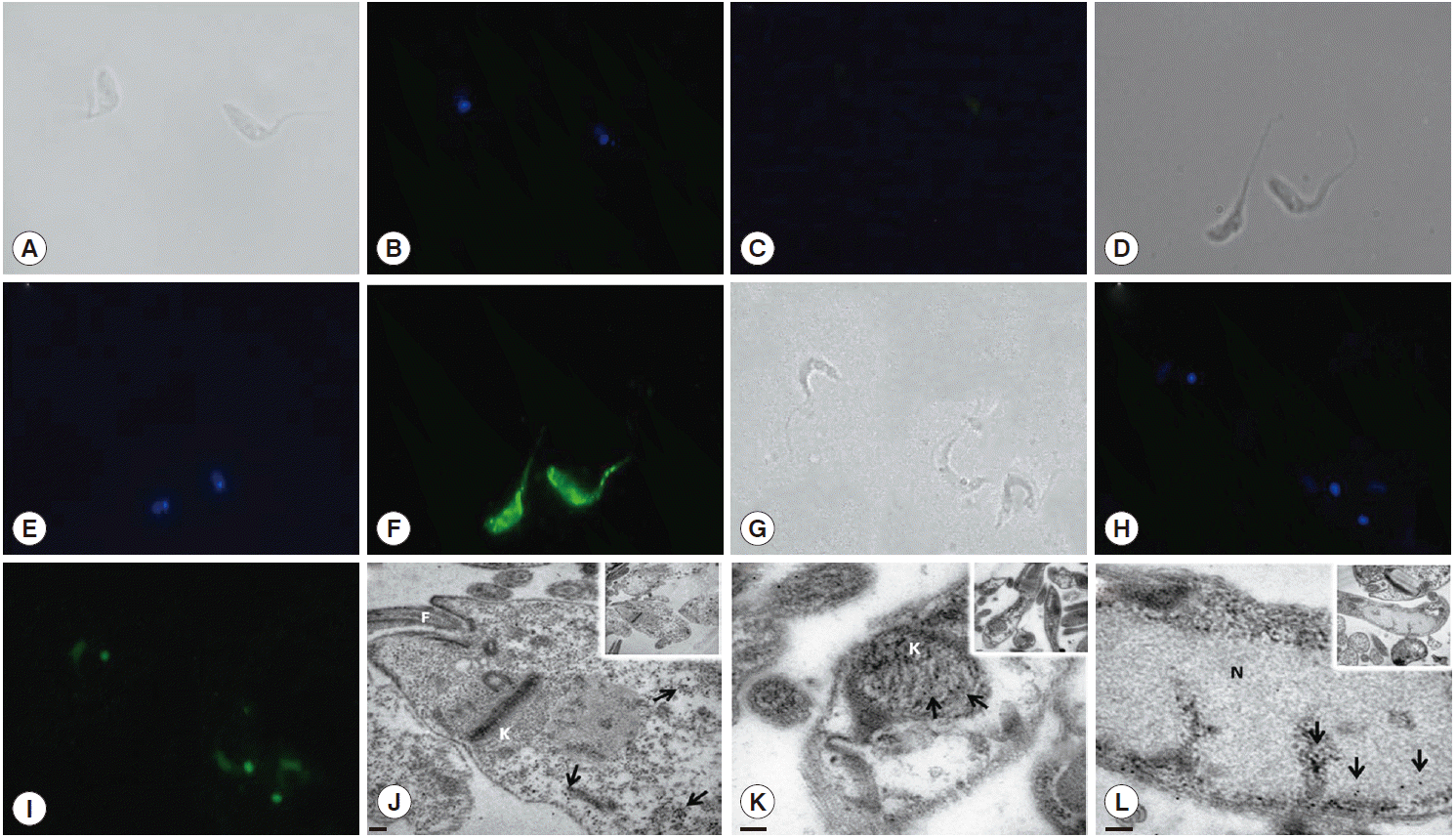
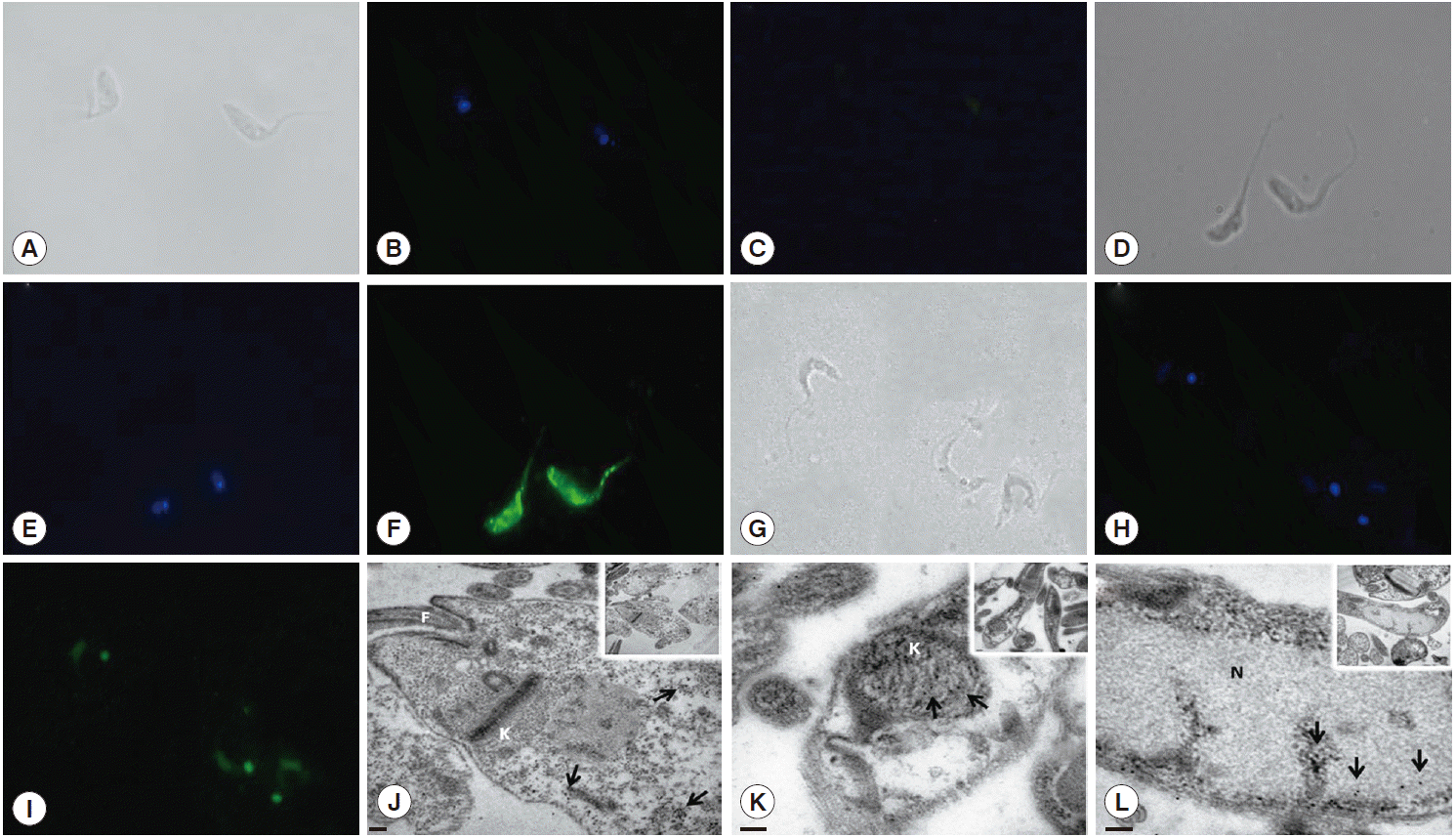
Fig. 2D). This differential recognition was also seen in the immunofluorescence images. Tc8.2 protein showed a cytoplasmic distribution in epimastigotes, and it was observed in the nucleus and kinetoplast in trypomastigotes. To verify the immunolocalization, a control using Tc8.2 protein adsorbed serum was used which showed that there was no labeling (
Fig. 3). These results were confirmed in the ultrastructural analysis, in which images showing gold particles were found in the nucleus and kinetoplast of trypomastigotes and in the cytoplasm of epimastigotes (
Fig. 3).
These differential characteristics could also be due to posttranslational modifications. Bioinformatics analyses indicated the presence of a transmembrane helix in the end portion of the amino acid sequence (position 564-586), the region corresponding to the hydrophobic motifs. In addition, there were glycosylation and phosphorylation recognition sites. For O-glycosylation, 10 possible sites were shown, while only 1 for N-glycosylation, corresponding to an “NMSI” amino acid sequence (0.6968 potential). With regard to phosphorylation events, the analysis showed the prediction of 8 Ser, 4 Thr and 2 Tyr residues, and protein kinase C (
Fig. 1A).
The function of
Tc8.2 is still unknown. The alignment of the nucleotide sequence of the gene in the Tritryp database has identified a copy in the
T. brucei genome (access no. Tb927.9.2650), which has been analyzed by parallel sequencing of RNA interference targets (RIT-seq) [
6], and the results suggested that blocking the gene is associated only with pronounced defects in blood forms of the parasite. In addition to these data, other characteristics of the gene have been examined by mRNA expression profile during the
T. brucei life cycle in in vivo assays [
7]. According to the results, the expression of the corresponding
Tc8.2 gene in
T. brucei AnTat1.1 is upregulated in stumpy forms compared to the other stages of the cycle. However, in the study performed with
T. brucei strain TREU927/4, the expression profiling of the life cycle stages showed no difference [
8]. In addition,
T. brucei Lister 427 transcriptome using blood and procyclic forms by Illumina method also showed similar levels of transcript in the 2 forms of the parasite [
9].
When the expression of the
Tc8.2 gene in
T. brucei Lister 427 was analyzed in response to environmental stress conditions like temperature variation by microarray [
10], RNA levels were practically not affected in the parasites incubated at high temperature, when compared with the control. Furthermore, the role of DHH1 helicase, which is required for RNA instability under certain conditions during the phase in the insect, was evaluated for gene expression. Using mutant models, it was observed that both the absence and overexpression of DHH1 did not affect
Tc8.2 RNA levels in
T. brucei [
10].
In
T. cruzi, transcripts analysis also showed that
Tc8.2 gene is expressed in all forms, corroborating to
T. brucei data, suggesting the importance of this protein during parasite life cycle. Interestingly, further
Tc8.2 characterization in
T. cruzi also showed different transcript sizes and location. Dual localization of
Tc8.2 could result from an alternative trans-splicing processing, as a consequence of the insertion of an mRNA leader sequence in different portions of the transcript 5´ end [
11]. This phenomenon has been reported in trypanosomatids, for example,
T. cruzi LYT1 [
12]. This protein is found both in the mitochondrion and in the plasma membrane of the parasite, and it has been suggested that they may have different functions depending on their location [
13,
14]. In addition, our results do not discard the possibility of post-translational processing, since there are several predicted sites of glycosylation and phosphorylation.
Overall, our findings showed that Tc8.2 has dual locations, but further analysis of the N-terminal sequence should be performed to search for splice acceptor sites, since our results indicated that this gene is alternatively expressed in epimastigote and trypomastigote forms. Additional studies are needed to determine the role of Tc8.2 protein in different forms of T. cruzi and if differences really have anything to do with the stage of development, where they can provide important information about the mechanism of gene expression in T. cruzi.
Notes
-
The authors declare no conflict of interest.
The authors thank the technicians Jussevania Pereira Santos and Ediel Clemente Costa for their help with laboratory maintenance and Vera Lúcia Hideko Tatakihara and Aparecida Donizette Malvezi for their help with T. cruzi maintenance. Dr. A. Leyva helped with English editing of this manuscript.
Fig. 1.Bioinformatics analysis of Tc8.2 gene. (A) Alignment of the amino acid sequences derived from the 2 copies of the Tc8.2 gene identified in the Trypanosoma cruzi genome (clone CL Brener) E-value = 0.0. The amino acid sequences (TcCLB.507615.100 and TcCLB.508347.80) were aligned with the ClustalW algorithm using the MegAlign program (DNASstar Inc.). The differences between the 2 sequences are highlighted in dark gray. Colored boxes indicate the sites to post translational modifications: transmembrane helix and hydrophobic motif (blue), O-glycosylation (yellow), N-glycosylation (green), phosphorylation events, Ser residues (red), Thr residues (purple), and Tyr residues (pink), and protein kinase C (brown). (B) Dendrogram of the amino acid sequences derived from the different copies of the Tc8.2 gene identified in the genome of different trypanosomatids. The amino acid sequences identified in T. cruzi clone CL Brener (TcCLB.507615.100 and TcCLB.508347.80), T. cruzi Sylvio (TcSYLVIO_003582) (E-value=0.0), T. cruzi marinkellei (Tc_MARK_2295) (E-value=0.0), Trypanosoma brucei (Tb927.9.2650) (E-value=7e-167), Leishmania donovani (LdBPK_261870) (E-value=4e-115), L. major (LmjF.26.1860) (E-value=5e-115), L. mexicana (LmxM.26.1860) (E-value=3e-118), and L. braziliensis (LbrM.26.1790) (E-value=1e-120). The construction was performed using the ClustalW and MEGA 6.06 program. The scale bar represents 0.05 nucleotide substitutions per site.
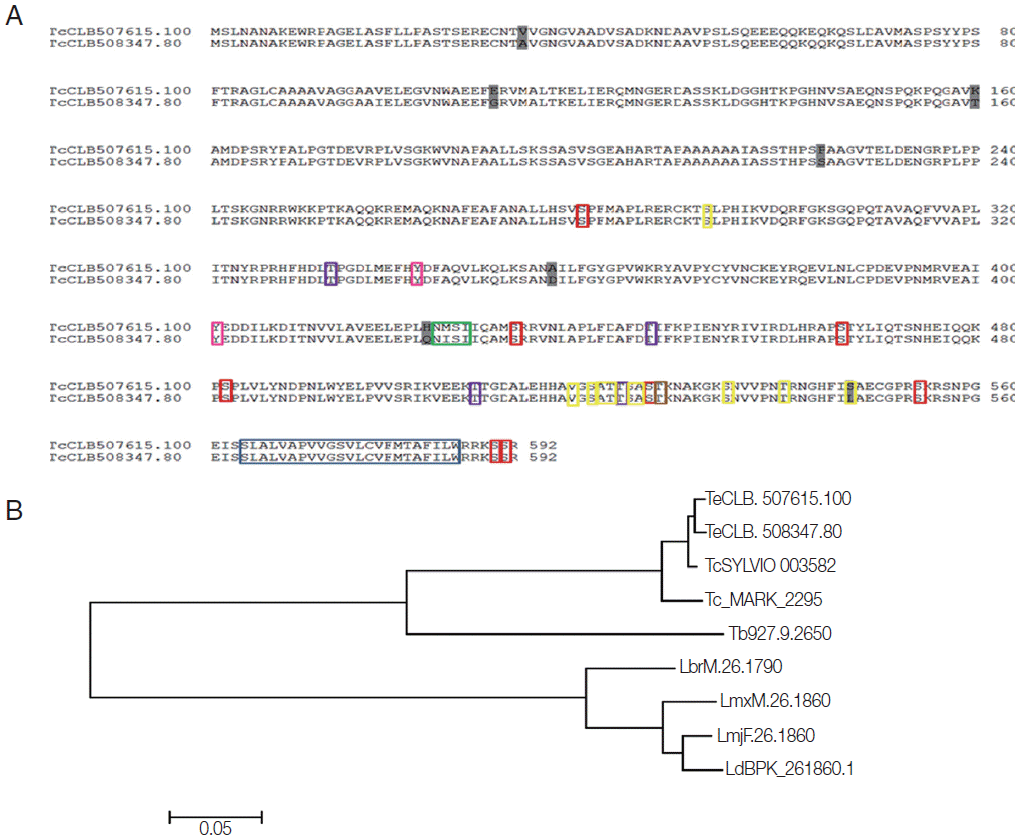
Fig. 2.Characterization of Tc8.2 gene. (A) Northern blot analysis of total RNA of epimastigotes (E), trypomastigotes (T) and amastigotes (A) of Trypanosoma cruzi and hybridization of 32P-labeled Tc8.2. MSM: molecular size marker. (B) RT-PCR of epimastigote and trypomastigote RNA. The cDNA was synthesized using oligo (dT), and PCR amplification was performed using specific primers for the Tc8.2 gene. Lanes: 1, genomic DNA of T. cruzi; 2-3, epimastigote cDNA; 4-5, trypomastigote cDNA synthetized with (RT+) and without (RT-) reverse transcriptase. (C) Expression of the Tc8.2 recombinant protein. Escherichia coli Bl21 (DE) was transformed with the clone pDEST/Tc8.2 and induction of protein obtained with 1 mM IPTG. Lanes: 1, non-induced culture; 2, IPTG-induced culture. (D) Protein extract of T. cruzi separated by 13% SDS-PAGE and transferred to a nitrocellulose membrane. Polyclonal antibodies against Tc8.2 protein recognize peptides in the extract of epimastigotes (E) and trypomastigotes (T).
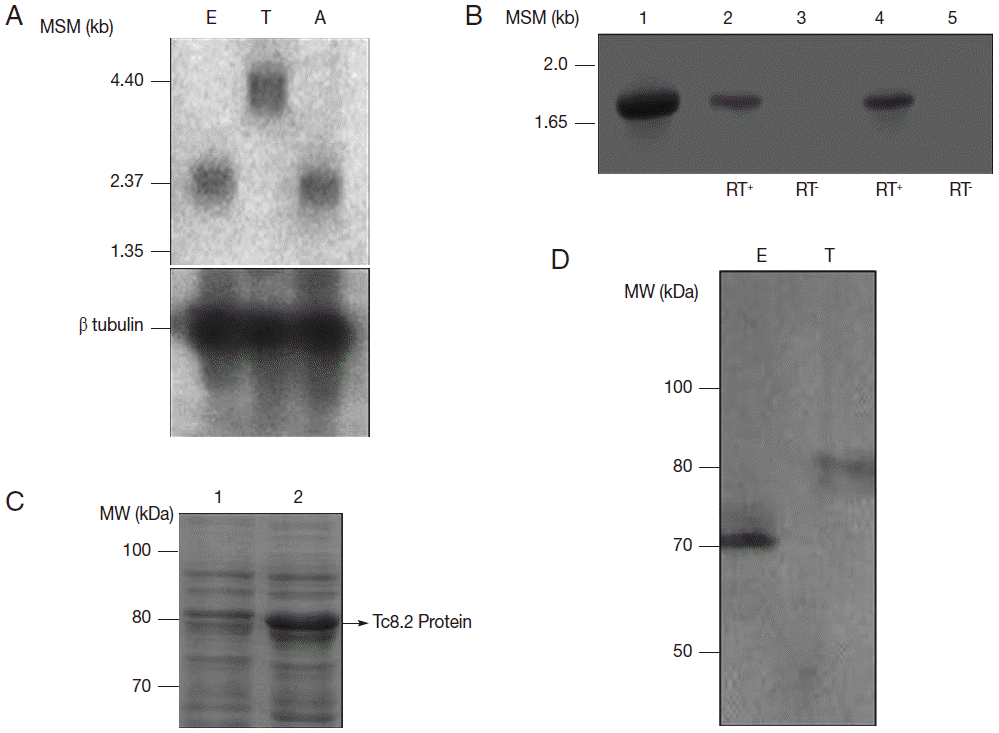
Fig. 3.Immunolocalization of Tc8.2 protein. Epimastigotes and trypomastigotes of Trypanosoma cruzi were fixed and permeabilized for immunofluorescence. Polyclonal antibodies against Tc8.2 protein recognize proteins differentially compartmentalized in T. cruzi. (A-C) Control of background fluorescence. (D-F) Epimastigotes. (G-I) Trypomastigotes. (A, D, and G) Differential interference contrast (DIC) showing the morphology of parasites. (B, E, and H) Cells stained with DAPI showing the nucleus and kinetoplast. (C) Control using serum protein adsorbed to Tc8.2. (F and I) Fluorescence of Alexa-Fluor 488 indicating Tc8.2 protein in the parasite. Ultrastructural detection of Tc8.2 protein, the distribution of gold particles in epimastigotes (J) and trypomastigotes (K and L). K, kinetoplast; N, nucleus; F, flagellum. Arrows show the gold particles. (J) Bar=0.2 μm. (K) Bar=100 nm. (L) Bar=100 nm.
References
- 1. World Health Organization. Chagas disease (American trypanosomiasis). WHO Technical Report Series. Geneva, Switzerland. WHO; 2012.
- 2. El Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, da Silveira JF, de Jong P, Edwards K, Englund PT, Fazelina G, Feldblyum T, Ferella M, Frasch AC, Gull K, Horn D, Hou L, Huang Y, Kindlund E, Klingbeil M, Kluge S, Koo H, Lacerda D, Levin MJ, Lorenzi H, Louie T, Machado CR, McCulloch R, McKenna A, Mizuno Y, Mottram JC, Nelson S, Ochaya S, Osoegawa K, Pai G, Parsons M, Pentony M, Pettersson U, Pop M, Ramirez JL, Rinta J, Robertson L, Salzberg SL, Sanchez DO, Seyler A, Sharma R, Shetty J, Simpson AJ, Sisk E, Tammi MT, Tarleton R, Teixeira S, Van Aken S, Vogt C, Ward PN, Wickstead B, Wortman J, White O, Fraser CM, Stuart KD, Andersson B. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 2005;309:409-415.
- 3. Medina-Acosta E, Cross GAM. Rapid isolation of DNA from trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol Biochem Parasitol 1993;59:327-329.
- 4. Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proc Natl Acad Sci USA 1979;76:4350-4354.
- 5. Porcile PE, Santos MR, Souza RT, Verbisck NV, Brandão A, Urmenyi T, Silva R, Rondinelli E, Lorenzi H, Levin MJ, Degrave W, Franco da Silveira J. A refined molecular karyotype for the reference strain of the Trypanosoma cruzi genome project (clone CL Brener) by assignment of chromosome markers. Gene 2003;308:53-65.
- 6. Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 2011;21:915-924.
- 7. Kabani S, Fenn K, Ross A, Ivens A, Smith TK, Ghazal P, Matthews K. Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics 2009;10:427.
- 8. Jensen BC, Sivam D, Kifer CT, Myler PJ, Parsons M. Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics 2009;10:482.
- 9. Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genomewide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res 2010;38:4946-4957.
- 10. Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J Cell Sci 2008;121:3002-3014.
- 11. Karniely S, Pines O. Single translation-dual destination: mechanism of dual protein targeting in eukaryotes. EMBO J 2005;6:420-425.
- 12. Benabdellah K, González-Rey E, González A. Alternative transsplicing of the Trypanosoma cruzi LYT1 gene transcript results in compartmental and functional switch for the encoded protein. Mol Microbiol 2007;65:1559-1567.
- 13. Manning-Cela R, Cortés A, Gonzáles-Rey E, Van Voorhis WC, Swindle J, González A. LYT1 protein is required for efficient in vitro infection by Trypanosoma cruzi. Infect Immun 2001;69:3916-3923.
- 14. Manning-Cela R, Gonzalez A, Swindle J. Alternative splicing of LYT1 transcripts in Trypanosoma cruzi. Infect Immun 2002;70:4726-4728.