Abstract
A study of 426 rabbits from 3 cities in Jilin province (Changchun City and Jilin City) and Liaoning province (Shenyang City) was conducted between May and June 2015. The overall prevalence of E. bieneusi in rabbits was 0.94% (4/426), with 0% (0/116), 1.72% (3/174), and 0.74% (1/136) in Jilin, Changchun, and Shenyang City, respectively. Only 3 farms (farm 1 and farm 3 in Changchun City, farm 8 in Shenyang City) were PCR-positive for E. bieneusi. Moreover, rabbits of more than 6 months (1.72%) had the highest E. bieneusi prevalence, followed by rabbits of 4-6 months (1.26%), 2-3 months (0.58%), and less than 1 month (0%). Analysis of ITS gene of E. bieneusi suggested that all 4 E. bieneusi isolates were genotype D, and were classified as group 1a. The present results first demonstrated the existence of zoonotic E. bieneusi in domestic rabbits in China. Effective control measures should be implemented to prevent E. bieneusi infection in domestic rabbits, other animals, and humans.
-
Key words: Encephalitozoon bieneusi, domestic rabbit, prevalence, internal transcribed spacer (ITS), China
China has a long history of breeding rabbits which have a close relationship with humans. With the improvement of living standards, rabbit meat became widely used for consumption. Thus, more people have pay attention to rabbit industry. Therefore, evaluating how pathogens influence animal health is essential to rabbit industry.
It is well known that the rabbit is a major host for many pathogens including microsporidia [
1,
2]. However, information regarding the prevalence and genotypes of
Encephalitozoon bieneusi in China is scarce. The objectives of the present study were to estimate the prevalence of
E. bieneusi in rabbits and to identify their genotypes in China. Moreover, the present study also aimed to provide the foundation data for further study of
E. bieneusi genotypes distribution in China.
A total of 426 fecal samples were collected from Jilin (n=290) and Liaoning (n=136) province between May and June 2015. At the time of sampling, all the rabbits were in good health. Fresh fecal sample was collected from each animal using sterile disposal PE gloves after defecation onto the ground. All samples were sent to the laboratory, and then genomic DNA was extracted using an EZNAR Stool DNA kit (OMEGA Biotec, Madison, Wisconsin, USA) according to the manufacturer instructions, immediately. All the extracted DNA samples were stored at -20˚C until PCR analysis. Information concerning gender, geographic origin, and age were obtained by a questionnaire.
E. bieneusi genotypes were determined by nested PCR of the internal transcribed spacer (ITS) region of the ribosomal RNA (rRNA) gene [
3]. PCR reaction (25 μl) was composed of 200 μM each deoxy-ribonucleoside triphosphate (dNTP), 1×Ex Taq buffer (Mg2
+ free), 0.4 μM of each primer, 0.625 U of Ex Taq DNA polymerase (TAKARA, Kyoto, Japan), 2 mM MgCl
2, and 2 μl of DNA template. The cycling conditions were denatured at 95˚C for 5 min, and then 35 cycles of 45 sec at 94˚C, 45 sec at 55˚C, and 1 min at 72˚C, followed by 72˚C for 10 min. Both positive and negative controls were included in each test. All the PCR products were electrophoresed on a 2% agarose gel, stained with GoldView™ (Solarbio, Beijing, China), and then observed under UV light.
Positive secondary PCR products were sent to Genscript Company (Nanjing, China) for sequencing. The obtained sequences were aligned with reported sequences of
E. bieneusi to determine the
E. bieneusi genotypes using the BLAST (
http://www.ncbi.nlm.nih.gov/BLAST/). Mega 5.0 software was used to construct the phylogenetic trees using neighbor-joining (NJ) method (Kimura 2-parameter model, 1,000 replicates).
Differences in the prevalence of E. bieneusi in rabbits of different geographical locations, gender groups, and age groups were analyzed by the chi-square test using SAS version 9.1 (SAS Institute Inc., Cary, North Carolina, USA). If P<0.05, the results were considered as statistically significant. Then, 95% confidence interval (CI) was provided in this study. Representative nucleotide sequences were deposited in GenBank with the accession no. KT325924.
A total of 4 (0.94%) out of 426 rabbit feces were
E. bieneusipositive, with 0% (0/116), 1.72% (3/174), and 0.74% (1/136) in Jilin, Changchun, and Shenyang City, respectively (
Table 1). Only 3 farms (farm 1 and farm 3 in Changchun, farm 8 in Shenyang) were PCR-positive for
E. bieneusi, with the highest infection rates in farm 1 (n=2, 3.2%), followed by farm 3 (n=1, 2.1%) and farm 8 (n=1, 1.8%) (
Table 1). Moreover, rabbits of more than 6 months (1.72%, 95% CI 0.00-5.07) revealed the highest
E. bieneusi prevalence, followed by rabbits of 4-6 months (1.26%, 95% CI 0.00-2.99), 2-3 months (0.58%, 95% CI 0.00-1.72), and less than 1 month (0%) (
Table 2). Analysis of ITS gene of
E. bieneusi revealed that all
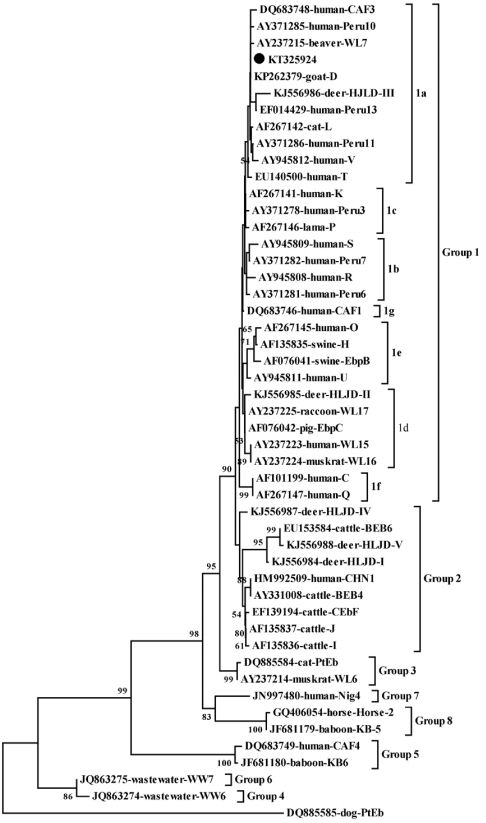
E. bieneusi isolates were genotype D (n=4), which belonged to group 1a (
Fig. 1).
To date, information concerning distribution of genotypes of
E. bieneusi in China is scarce, especially in rabbits. In order to further improve the prevalence and distribution information, we conducted the present study. The overall prevalence of
E. bieneusi infection in rabbits was 0.94%, which was considerably lower than that in other animals in northeastern China (
Table 3). The lower infection rates may be related to different susceptibility of different animals, sample sizes and geo-ecological conditions, standardized management mode, and so on. Moreover, the prevalence of
E. bieneusi in rabbits seemed to increase with the age in the present study, which agreed to previous studies;
E. bieneusi was an opportunistic parasite and it may accumulate throughout the life time [
4,
5]. Moreover, probably because of similar climates in Changchun City, Jilin City, and Shenyang City, and the same management mode in these farms, the positive rates of
E. bieneusi infection in rabbits among the 9 farms in northern China were not statistically different (
P>0.05).
In the present study, only genotype D (n=3) was found in rabbits. This result was similar to a previous study conducted in Spain, in which also only genotype D was identified [
6]. Genotype D not only has a wide geographical distribution but also can infect almost all host species, including humans, domestic animals, and wild animals [
6]. In China, genotype D also has a broad host range. Besides some animals, e.g., golden takins (
Budorcas taxicolor bedfordi) and panda in Shanxi [
7,
8], pet chinchillas in Anyang, Beijing, and Zhengzhou, China [
9], nonhuman primates in Henan, Guangxi, Guangdong, Yunnan [
10], and Sichuan [
11], dairy cattle, goats, sheep, pigs, cats, and dogs in northern China [
12-
15]. Genotype D infection was also reported in children in Shanghai [
16] and HIV patients in Henan [
17], which suggest that
E. bieneusi genotype D could readily transmit between humans and animals. More importantly, genotype D was also present in wastewater which may lead to outbreaks of this disease [
18]. Rabbits may play an important role and cannot be ignored during this pathogen infection process in China. Moreover, sequence analysis of the ITS rRNA gene showed that the obtained sequences were identical to the reference sequence from sheep and goat isolates (accession no. KP262379) in China, which revealed that the infectious source is more likely to be excreted from sheep and goats. Therefore, we should pay serious attention to nosocomial transmission among different animals.
In view of such situation, the following practice should be performed to prevent transmission of E. bieneusi among rabbits, other animals, and humans in China: first, different kinds of animals should be managed strictly separately; second, the prevalence of E. bieneusi should be examined periodically; third, feces should be cleaned timely; fourth, parasites should be expelled regularly; fifth, humans should have a habit of wash hands before meals and after contact with animals.
In conclusion, the present study first demonstrated existence (0.94%, 4/426) of E. bieneusi infection in rabbits in China, and the infection rate was increased with age. Moreover, only genotype D was identified in rabbits which should be considered as an important potential source for E. bieneusi infection in China. Therefore, effective strategies should be taken to prevent and control of E. bieneusi infection in rabbits, other animals, and humans in China.
Notes
-
The authors declare that they have no competing interests.
This work was supported by the National High-Tech R&D Program of China (863 Programs) (no. 2013AA102806, no. 2011AA10A215), National Natural Science Foundation of China (no. 31272552, no. 31272541), the Key Scientific and Technological Project of Jilin Province (no. 20140204068NY), Jilin Province Science and Technology Development Program of China (no. 20111816), and Jilin Province Quality and Safety of Agricultural Products Program by World Bank (no. 2011-Y07).
Fig. 1.Phylogenetic analysis of Enterocytozoon bieneusi using Neighbor-Joining (NJ) method based on sequences of the internal transcribed spacer (ITS) region. The numbers at notes indicate bootstrap values (1,000 replicates, values below 50% are not shown). E. bieneusi isolate identified in the present study is indicated by solid circle.
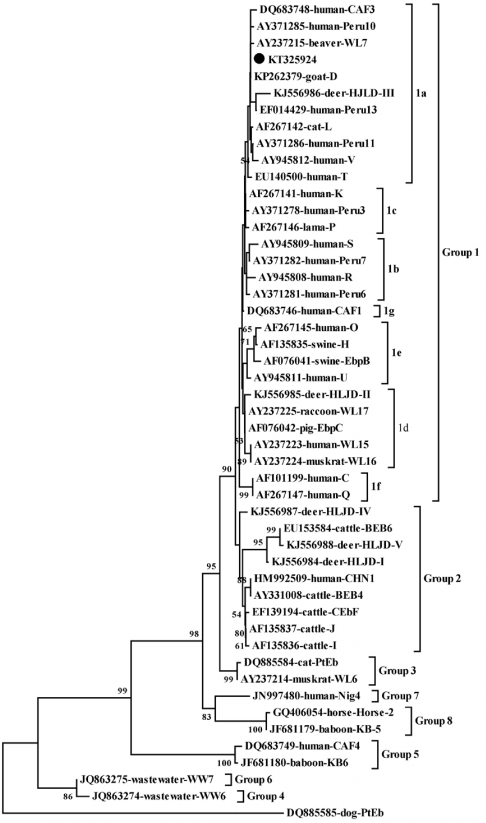
Table 1.
Enterocytozoon bieneusi genotypes identified in rabbits in different farms
Table 1.
|
Region |
Farm ID |
Age category (n) |
No. positive/no. tested (%) |
Genotype (n) |
|
Changchun City |
Farm 1 |
< 1 month (15), 2-3 months (21), 4-6 months (26) |
2/62 (3.2) |
D (n = 2) |
|
Farm 2 |
> 6 months (33) |
0/33 (0) |
- |
|
Farm 3 |
< 1 month (11), 2-3 months (16), 4-6 months (20) |
1/47 (2.1) |
D (n = 1) |
|
Farm 4 |
4-6 months (32) |
0/32 (0) |
- |
|
Jilin City |
Farm 5 |
2-3 months (13), 4-6 months (50), > 6 months (14) |
0/77 (0) |
- |
|
Farm 6 |
< 1 month (11), 2-3 months (28) |
0/39 (0) |
- |
|
Shenyang City |
Farm 7 |
2-3 months (21), 4-6 months (15) |
0/36 (0) |
- |
|
Farm 8 |
2-3 months (41), 4-6 months (16) |
1/57 (1.8) |
D (n = 1) |
|
Farm 9 |
2-3 months (32), > 6 months (11) |
0/43 (0) |
- |
|
Total |
|
< 1 month (37), 2-3 months (172), 4-6 months (159), > 6 months (58) |
3/426 (0.94) |
D (n = 4) |
Table 2.
Enterocytozoon bieneusi genotypes identified in rabbits in different age groups
Table 2.
|
Factor |
Category |
No. tested |
No. positive |
% (95% CI) |
Genotype (n) |
|
Age |
< 1 month |
37 |
0 |
0 (-) |
- |
|
2-3 months |
172 |
1 |
0.58 (0.00-1.72) |
D (n = 1) |
|
4-6 months |
159 |
2 |
1.26 (0.00-2.99) |
D (n = 2) |
|
> 6 months |
58 |
1 |
1.72 (0.00-5.07) |
D (n = 1) |
Table 3.
Enterocytozoon bieneusi genotypes identified in different animals in northeastern China
Table 3.
|
Country |
Hosts |
No. tested |
% |
Microsporidia species |
Reference |
|
Heilongjiang and Jilin |
Cattle |
537 |
6.0 |
BEB4, BEB6, CM7, CS-4, EbpC, G, I, J, OEB1, and 11 new genotypes (NESH1 to NESH6 and NECA1 to NECA5) |
[19] |
|
Sheep |
498 |
13.9 |
|
|
Heilongjiang |
Dairy cattle |
133 |
30.1 |
O, EbpA, I, J, D, BEB4, and three novel genotypes (CC-I to CC-III) |
[14] |
|
Heilongjiang |
Sheep |
138 |
22.5 |
Peru6, BeB6, D, EbpC, EbpA, COG-I |
[13] |
|
Heilongjiang |
Goats |
55 |
21.8 |
BeB6, Peru6, D, O, COS-I to COS-VII |
|
|
Heilongjiang |
Cervus nippon
|
52 |
25.0 |
BeB6, HLJD-I–HLJD-V |
|
|
Heilongjiang |
Cervus elaphus
|
5 |
20.0 |
HLJD-V |
[4] |
|
Jilin |
Cervus nippon
|
34 |
44.1 |
BeB6, HLJD-V |
|
|
Jilin |
Pigs |
156 |
60.3 |
CHN7, CS-1, CS-4, CS-6, EbpA, EbpB, EbpC, EbpD, EBITS3, G, and Henan-I; one new: CS-9 |
[20] |
|
Heilongjaing |
Sheep |
45 |
4.4 |
BEB6 |
|
|
Heilongjiang |
Pigs |
95 |
89.5 |
H, HLJ-I, HLJ-II, HLJ-III, HLJ-IV, CS-1, O, LW1, EbpA, D |
[15] |
|
Heilongjiang |
Cats |
52 |
5.8 |
Human pathogenic genotypes D and IV |
[12] |
|
Heilongjiang |
Dogs |
267 |
6.7 |
D, EbpC, NED1, NED2, NED3, NED4, and PtEb IX |
|
|
Heilongjiang, Jilin, Inner Mongolia |
Pigs |
113 |
45.1 |
D, EbpA, EbpC, EbpD, H, Henan-I, Henan-III, Henan-IV, and O, and eight new genotypes (CS-1 to CS-8) |
[21] |
|
Jilin and Liaoning |
Rabbits |
426 |
0.94 |
D |
This study |
References
- 1. Shin JC, Kim DG, Kim SH, Kim S, Song KH. Seroprevalence of Encephalitozoon cuniculi in pet rabbits in Korea. Korean J Parasitol 2014;52:321-323.
- 2. Yang D, Ren Y, Fu Y, Xie Y, Nie H, Nong X, Gu X, Wang S, Peng X, Yang G. Genetic variation of Taenia pisiformis collected from Sichuan, China, based on the mitochondrial cytochrome B gene. Korean J Parasitol 2013;51:449-452.
- 3. Hu Y, Feng Y, Huang C, Xiao L. Occurrence, source, and human infection potential of Cryptosporidium and Enterocytozoon bieneusi in drinking source water in Shanghai, China, during a pig carcass disposal incident. Environ Sci Technol 2014;48:14219-14227.
- 4. Zhao W, Zhang W, Wang R, Liu W, Liu A, Yang D, Yang F, Karim MR, Zhang L. Enterocytozoon bieneusi in sika deer (Cervus nippon) and red deer (Cervus elaphus): deer specificity and zoonotic potential of ITS genotypes. Parasitol Res 2014;113:4243-4250.
- 5. Thellier M, Breton J. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite 2008;15:349-358.
- 6. Galván-Díaz AL, Magnet A, Fenoy S, Henriques-Gil N, Haro M, Gordo FP, Miró G, del Águila C, Izquierdo F. Microsporidia detection and genotyping study of human pathogenic E. bieneusi in animals from Spain. PLoS One 2014;9:e92289.
- 7. Tian GR, Zhao GH, Du SZ, Hu XF, Wang HB, Zhang LX, Yu SK. First report of Enterocytozoon bieneusi from giant pandas (Ailuropoda melanoleuca) and red pandas (Ailurus fulgens) in China. Infect Genet Evol 2015;34:32-35.
- 8. Zhao GH, Du SZ, Wang HB, Hu XF, Deng MJ, Yu SK, Zhang LX, Zhu XQ. First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi). Infect Genet Evol 2015;34:394-401.
- 9. Qi M, Luo N, Wang H, Yu F, Wang R, Huang J, Zhang L. Zoonotic Cryptosporidium spp. and Enterocytozoon bieneusi in pet chinchillas (Chinchilla lanigera) in China. Parasitol Int 2015;64:339-341.
- 10. Karim MR, Wang R, Dong H, Zhang L, Li J, Zhang S, Rume FI, Qi M, Jian F, Sun M, Yang G, Zou F, Ning C, Xiao L. Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Appl Environ Microbiol 2014;80:1893-1898.
- 11. Ye J, Xiao L, Li J, Huang W, Amer SE, Guo Y, Roellig D, Feng Y. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol Int 2014;63:132-137.
- 12. Li W, Li Y, Song M, Lu Y, Yang J, Tao W, Jiang Y, Wan Q, Zhang S, Xiao L. Prevalence and genetic characteristics of Cryptosporidium, Enterocytozoon bieneusi and Giardia duodenalis in cats and dogs in Heilongjiang province, China. Vet Parasitol 2015;208:125-134.
- 13. Zhao W, Zhang W, Yang D, Zhang L, Wang R, Liu A. Prevalence of Enterocytozoon bieneusi and genetic diversity of ITS genotypes in sheep and goats in China. Infect Genet Evol 2015;32:265-270.
- 14. Zhao W, Zhang W, Yang F, Zhang L, Wang R, Cao J, Shen Y, Liu A. Enterocytozoon bieneusi in Dairy Cattle in the Northeast of China: Genetic Diversity of ITS Gene and Evaluation of Zoonotic Transmission Potential. J Eukaryot Microbiol 2015;62:553-560.
- 15. Zhao W, Zhang W, Yang F, Cao J, Liu H, Yang D, Shen Y, Liu A. High prevalence of Enterocytozoon bieneusi in asymptomatic pigs and assessment of zoonotic risk at the genotype level. Appl Environ Microbiol 2014;80:3699-3707.
- 16. Wang L, Xiao L, Duan L, Ye J, Guo Y, Guo M, Liu L, Feng Y. Concurrent infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Negl Trop Dis 2013;7:e2437.
- 17. Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, Liu L, Feng Y, Xiao L. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol 2013;51:557-563.
- 18. Guo Y, Alderisio KA, Yang W, Cama V, Feng Y, Xiao L. Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Appl Environ Microbiol 2014;80:218-225.
- 19. Jiang Y, Tao W, Wan Q, Li Q, Yang Y, Lin Y, Zhang S, Li W. Erratum for Jiang et al., Zoonotic and potentially host-adapted Enterocytozoon bieneusi genotypes in sheep andcattle in northeast China and an increasing concern about the zoonotic importance of previously considered ruminant-adapted genotypes. Appl Environ Miicrobiol 2015;81:5278.
- 20. Li W, Li Y, Li W, Yang J, Song M, Diao R, Jia H, Lu Y, Zheng J, Zhang X, Xiao L. Genotypes of Enterocytozoon bieneusi in livestock in China: high prevalence and zoonotic potential. PLoS One 2014;9:e97623.
- 21. Li W, Diao R, Yang J, Xiao L, Lu Y, Li Y, Song M. High diversity of human-pathogenic Enterocytozoon bieneusi genotypes in swine in northeast China. Parasitol Res 2014;113:1147-1153.