Abstract
Giardia lamblia is a protozoan that causes diarrheal diseases in humans. Cytoskeletal structures of Giardia trophozoites must be finely reorganized during cell division. To identify Giardia proteins which interact with microtubules (MTs), Giardia lysates were incubated with in vitro-polymerized MTs and then precipitated by ultracentifugation. A hypothetical protein (GL50803_8405) was identified in the precipitated fraction with polymerized MTs and was named GlMBP1 (G. lamblia microtubule-binding protein 1). Interaction of GlMBP1 with MTs was confirmed by MT binding assays using recombinant GlMBP1 (rGlMBP1). In vivo expression of GlMBP1 was shown by a real-time PCR and western blot analysis using anti-rGlMBP1 antibodies. Transgenic G. lamblia trophozoites were constructed by integrating a chimeric gene encoding hemagglutinin (HA)-tagged GlMBP1 into a Giardia chromosome. Immunofluorescence assays of this transgenic G. lamblia, using anti-HA antibodies, revealed that GlMBP1 mainly localized at the basal bodies, axonemes, and median bodies of G. lamblia trophozoites. This result indicates that GlMBP1 is a component of the G. lamblia cytoskeleton.
-
Key words: Giardia lamblia, microtubule-binding protein 1
INTRODUCTION
Giardia lamblia infects humans as cysts that are converted into trophozoites, which multiply by binary fission in human intestines and cause diarrheal disease. A trophozoite of
G. lamblia has 2 nuclei and characteristic cytoskeletal structures such as a ventral disc, a median body, 4 pairs of flagella, and a funis [
1]. Positioning of these structures in the dividing
Giardia cells must be finely coordinated for successful proliferation. In eukaryotic organisms, microtubules (MTs) play an essential role in the coordinated movement of cellular structures by maintaining equilibrium between polymerization and depolymerization [
2]. Growing and shortening of MTs is mediated by MT-associated proteins, including end-binding 1 (EB1), which is a plus-end tracking protein [
3].
An EB1 homologous protein (GlEB1) was found in the flagellar tips, median bodies, and mitotic spindles of
G. lamblia [
4,
5]. The role of GlEB1 was assessed by complementation assays using a
BIM1 mutant of
Saccharomyces cerevisiae, in which proper positioning of the nucleus was abolished [
6]. Biochemical characterization of GlEB1 was performed to define the domains responsible for MT binding and dimerization [
5].
Investigations on the
Giardia cytoskeleton have focused on its unique structures such as the ventral disc and median body. Tubulin and
Giardia-specific giardin proteins were identified, via Triton X-100 extraction, as the main components of
G. lamblia ventral disc [
7]. Recent technical progress in proteomic analysis has led to the discovery of additional proteins associated with the ventral disc, whose function is yet to be defined [
8]. In addition, shotgun proteomics along with GFP-tagging of the purified ventral disc of
G. lamblia facilitated the identification of 18 novel disc-associated proteins [
9]. One of these disc-associated proteins, DAP116343, was also found in the median body and knockdown of this protein by morpholinos resulted in aberrant disc formation in
G. lamblia [
10].
Thus, dynamic MTs are expected to mediate cell division in G. lamblia. Various MT-binding proteins may play roles in this process. In this study, a novel MT-binding protein was discovered in Giardia lysates, using in vitro-polymerized MTs.
MATERIALS AND METHODS
Giardia cell culture and preparation of Giardia extracts
Trophozoites of the
G. lamblia WB strain (ATCC30957; American Type Culture Collection, Manassas, Virginia, USA) were grown for 72 hr in TYI-S-33 medium (2% casein digest, 1% yeast extract, 1% glucose, 0.2% NaCl, 0.2% L-cysteine, 0.02% ascorbic acid, 0.2% K
2HPO
4, 0.06% KH
2PO
4, 10% calf serum, and 0.5 mg/ml bovine bile, pH 7.1) [
11]. They were then resuspended in PBS (137 mM NaCl, 2.7 mM KCl, 10.1 mM Na
2HPO
4, and 2 mM KH
2PO
4, pH 7.4), and lysed by sonication.
The binding of Giardia lysates to polymerized MTs was performed in vitro using the Microtubule-Binding Protein Spin-Down Assay Kit BK029 (Cytoskeleton, Denver, Colorado, USA). MTs were assembled from 100 µg of pure tubulin (isolated from bovine brain; Cytoskeleton) in 20 µl of PEM [80 mM piperazine-N,N’-bis(2-ethanesulfonic acid), pH 6.8, 1 mM EGTA, and 1 mM MgCl2] in the presence of 1 mM GTP and 5% glycerol at 35˚C for 20 min, and immediately stabilized in 200 µl of warm PEM-20 µM taxol (Cytoskeleton). Twenty µmoles of the MTs were incubated with 100 µg of Giardia lysate in a total volume of 50 µl at 25˚C for 40 min. The reaction mixtures were then centrifuged with a 50% glycerol cushion-PEM-taxol mixture, at 100,000 g at 25˚C for 40 min in an ultracentrifuge (Hitachi Koki, Tokyo, Japan). The resulting pellet fraction was then resolved on an 8% polyacrylamide gel and visualized by silver staining. The same amount of Giardia extract was precipitated by ultracentrifugation, and compared side-by-side with the extracts precipitated with MTs.
Liquid chromatography mass spectrometry
The protein band present in the MT fraction was excised and digested with trypsin. The trypsin-treated proteins were analyzed by quadrupole time-of-flight (Q-TOF) mass spectrometry (MS) in addition to matrix-assisted laser desorption ionization-TOF MS (MALDI-TOF MS). Product ion spectra were collected in the information-dependent acquisition mode and were analyzed with an Agilent 6530 accurate-mass Q-TOF MS. For the Q-TOF liquid chromatography-tandem MS (LC-MS/MS) data sets, tandem mass spectra were submitted to our MASCOT inhouse database search engine (NCBI NR database downloaded on 31 July 2009). For protein identification, a MASCOT ion score of >37 was used as the criterion for a meaningful result.
Expression and purification of recombinant GlMBP1 (rGlMBP1)
A 1,338 bp DNA fragment encoding the GlMBP1 open reading frame (ORF) was amplified by PCR from the genomic DNA of
G. lamblia, using 2 primers, 8405F and 8405R (
Table 1), and cloned into pET21b (Novagen, Darmatadt, Germany), resulting in pETGlMBP1. The rGlMBP1 was expressed in Escherichia coli BL21 (DE3), with 0.5 mM ITPG at 30˚C for 3 hr, and purified using TALON Metal affinity chromatography, as described by the manufacturer (Clontech, Mountain View, California, USA).
The level of GlMBP1 mRNA expression was evaluated by real-time PCR. Total RNA was isolated from
G. lamblia, using TRIzol (Invitrogen, Carlsbad, California, USA). cDNA was synthesized from 5 µg of RNA using the ImProm-II
TM RT system (Promega, Madison, Wisconsin, USA) following the manufacturer’s directions. cDNA was then analyzed in the Light Cycler 480 II Real-Time PCR System (Roche Applied Science, Mannheim, Germany) using LightCycler 490 DNA SYBR Green I Master (Roche Applied Science). Conditions for real-time PCR were as follows: pre-incubation at 95˚C for 5 min followed by 45 amplification cycles of 95˚C for 10 sec, 56˚C for 20 sec, and 72˚C for 10 sec. Real-time PCR was carried out in triplicate in a 96-well plate using the specific primers listed in
Table 1. The tim gene encoding triose-1-phosphate isomerase of
G. lamblia was used as an endogenous control for the reactions.
Histidine-tagged rGlMBP1 was expressed in
E. coli BL21 (DE3) with the addition of 0.5 mM IPTG at 37˚C for 3 hr. The rGlMBP1 protein was excised from the acrylamide gel, and used to immunize Sprague-Dawley rats (2-week-old, female) to produce polyclonal antibodies as described previously [
12].
Ten µg of purified rGlMBP1 reacted with 20 µM polymerized MTs was used for MT-binding assays. After ultracentrifugation, the resulting soluble and pellet fractions were separated on 12% acrylamide gel, and then transferred to nitrocellulose membranes (Milipore, Bedford, Massachusetts, USA). The membrane was incubated with anti-histidine antibodies (1:5,000 dilution; IG Therapy, Chunchon, Korea) in a blocking solution (Tris-buffered saline containing 5% skim milk, and 0.1% Tween 20) at 4˚C overnight. Following incubation with alkaline phosphatase-conjugated anti-mouse IgG (1:2,000 dilution; Sigma, St. Louis, Missouri, USA), the immunoreactive protein was visualized in a nitro blue tetrazolium/5ʹ-bromo-4-chloro-3-indolyphosphate system (Promega).
Integration of HA-tagged glmbp1 into a Giardia chromosome
To tag a hemagglutinin (HA) epitope to the C-terminal portion of the
glmbp1 gene, a 1,538 bp DNA fragment containing the promoter (200 bp) and ORF of the
glmbp1 gene was amplified from
G. lamblia WB genomic DNA by PCR, using 2 primers, GlMBP-F and GlMBP-HA-R (
Table 1).
NcoI and
NotI sites, located at the ends of the resulting
glmbp1 DNA, were used for cloning into the corresponding sites of plasmid pGFP. pac [
13], resulting in the plasmid pGlMBP1HA.pac. This construct was verified by DNA sequencing (Macrogen, Seoul, Korea). The plasmid was linearized using its unique
PstI site, and 10 μg of linear DNA was then introduced into
Giardia trophozoites. Briefly, the trophozoites were grown for 72 hr in normal TYI-S-33 medium. Transfection of linearized DNA into
G. lamblia trophozoites was performed by electroporation under the following conditions: 350 volts, 1000 µF, and 700 Ω (BioRad, Hercules, California, USA). Trophozoites harboring pGlMBP1HA.pac were selected by adding puromycin (AG Scientific, San Diego, California, USA) to the TYI-S-33 medium at a final concentration of 10 µg/ml. After 7 or 8 days of cultivation, resistant cells were recovered. To confirm that the construct was integrated into the
Giardia genome in the transfected cells, PCR was performed for the isolated genomic DNA from pGlMBP1HA. pac-transfected cells, using a common 5ʹ-primer (GlMBP-F) and two 3ʹ-primers (GlMBP-HA-R or pac down) (
Table 1). PCR conditions were as follows: pre-denaturation at 94˚C for 5 min followed by 35 cycles of 94˚C for 1 min, 56˚C for 1 min, and 72˚C for 1 min 30 sec, and post-elongation at 72˚C for 10 min using a Veriti Thermal Cycler (Life Technologies, Foster City, California, USA).
To determine whether HA-tagged GlMBP1 was expressed in the transfected Giardia, cell extracts were prepared from G. lamblia WB and puromycin-resistant G. lamblia, and then were analyzed by sodium dodecyl-polyacrylamide gel electrophoresis (SDS-PAGE). The resulting membrane was incubated with rat anti-GlMBP1 antibodies (1:1,000), followed by incubation with horseradish peroxidase-conjugated anti-rat-IgG antibodies (1:1,000). Immunoreactive proteins were visualized using an Enhanced Chemiluminescence System (Youngin Frontier Company, Seoul, Korea). The membrane was incubated in a stripping buffer (ATTO Corporation, Tokyo, Japan) at room temperature for 30 min, and reacted with polyclonal G. lamblia anti-α-tubulin antibodies (1:10,000) as a loading control.
Immunofluorescence assay (IFA)
To examine the localization of GlMBP1 in G. lamblia expressing HA-tagged GlMBP1, the cells were attached to glass slides coated with L-lysine in a humidified chamber. The attached cells were fixed with chilled 100% methanol at -20˚C for 10 min and then permeabilized with PBS/0.5% Triton X-100 for 10 min. After 1-hr incubation in blocking buffer [PBS, 5% goat serum, and 3% bovine serum albumin (BSA)], the cells were reacted overnight with mouse anti-HA antibodies (1:50; Sigma). Following 3 times 5-min washes with PBS, the cells were incubated with AlexaFluor 488-conjugated anti-mouse IgG (1:200; Molecular Probes, Grand Island, New York, USA) at 37˚C for 1 hr. Cells were mounted on slides with VECTASHIELD Anti-fade Mounting Medium with 4ʹ,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, California, USA) and then were observed with an Axiovert 200 fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
RESULTS
Isolation of a G. lamblia MT-binding protein 1, GlMBP1
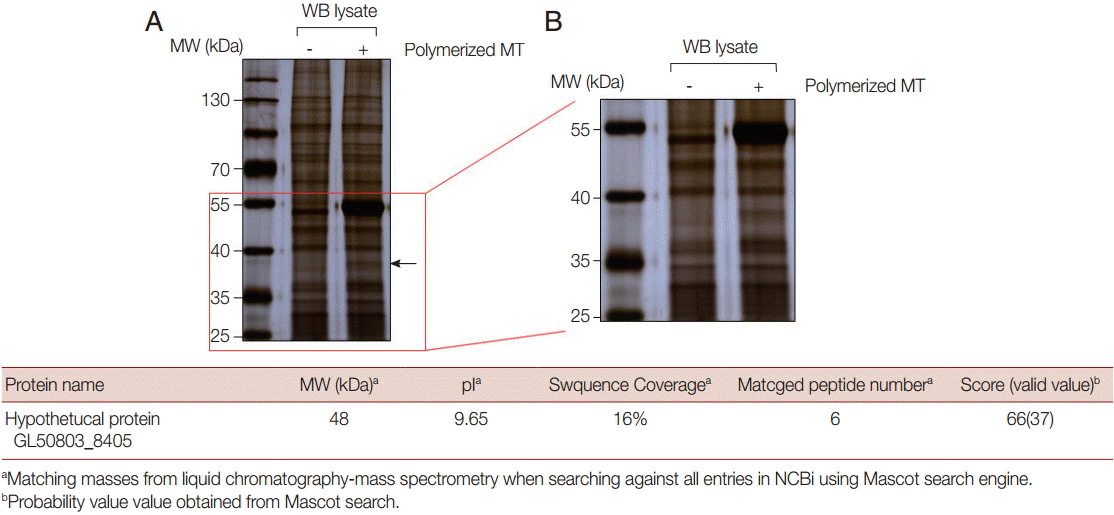
The pattern seen from proteins precipitated with MTs was almost identical to that of the control, except for 2 protein bands (
Fig. 1A). The larger and more abundant of these proteins was 55 kDa in size and was believed to be the tubulin monomer used in the MT-binding assay, while the smaller and less abundant protein was thought to be an MT-binding protein derived from
Giardia extracts. An extended view of the separated proteins between 55 kDa and 25 kDa revealed a protein of <40 kDa that was among the proteins precipitated with MTs, but was absent in the control proteins as indicated with an arrow (
Fig. 1B).
This protein was excised and processed for Q-TOF LC MS/MS analysis (
Fig. 1C). It was identified as a hypothetical protein, annotated as GL50803_8405, and named
G. lamblia MT-binding protein 1, GlMBP1. The putative protein had a molecular weight of 48 kDa and a pI value of 9.65. The Q-TOF LC-MS/MS analysis indicated 6 matched peptides, 16% coverage, and a meaningful MASCOT score of 66. Interestingly, the putative size of this annotated protein was 48 kDa whereas the excised protein was smaller (<40 kDa). This result suggested the possibility that the excised protein was a degradation product of GlMBP1.
The open reading frame (ORF) encoding GlMBP1 was made of 445 amino acids. A BlastP search of the protein database of other metazoan organisms yielded only homologous protein except a homologous protein (SS50377_11087) in Spironucleus salmonicida, a protozoan pathogen in salmons. A domain search of GlMBP1, as well as the homologous protein in S. salmonicida, did not show any MT-binding domains such as calponin homology, cytoskeletal associated protein-glycine rich, and tumor overexpressed gene domains, which are conserved in mammalian and yeasts. The only domains found in these proteins were C2HC-type zinc-finger domains.
Confirmation of the MT-binding ability of GlMBP1
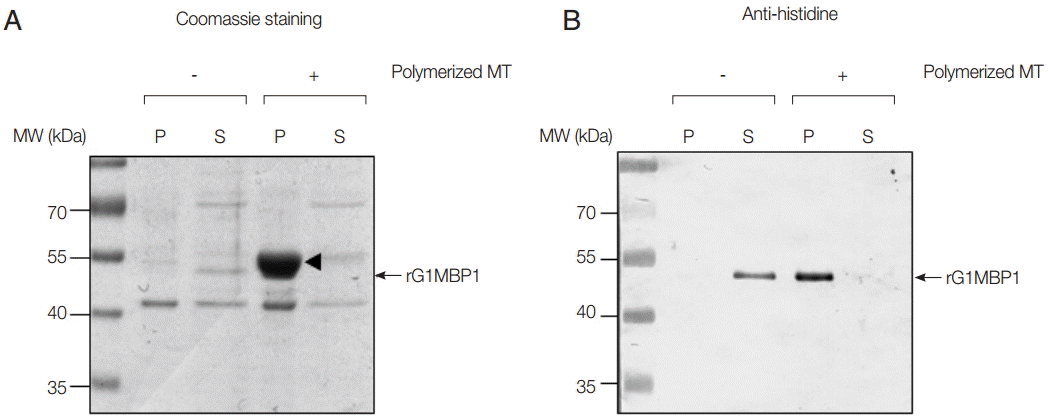
To confirm the association between GlMBP1 and MTs, an MT-binding assay was performed with recombinant GlMBP1 (rGlMBP1) and in vitro polymerized MTs. In the absence of MTs, histidine-tagged rGlMBP1 was found in the soluble fraction by Coomassie-staining and western blot analysis using anti-histidine antibodies (
Fig. 2A,
B, respectively). Upon incubation with polymerized MTs, rGlMBP1 was found in the pellet fraction along with tubulin monomer (
Fig. 2A). Because the sizes of tubulin and rGlMBP1 were similar, it was difficult to distinguish one from the other in the SDS-PAGE gel. However, western blotting using anti-histidine antibodies clearly showed that rGlMBP1 precipitated with polymerized MTs, indicating that rGlMBP1 binds to MTs (
Fig. 2B).
Even though GlMBP1 was identified as an MT-binding protein in
G. lamblia extracts, whether this protein is expressed in vivo in
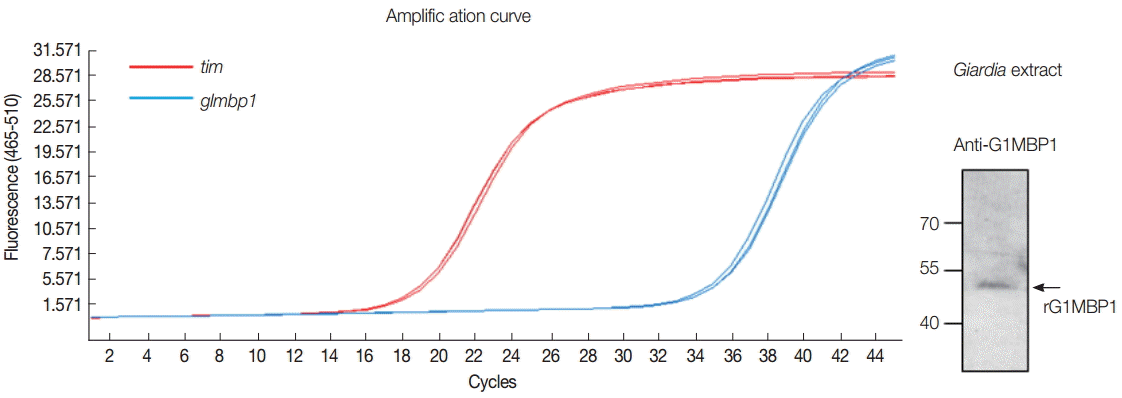
G. lamblia still needed to be determined. To assess the expression of
glmbp1 in
Giardia, the level of
glmbp1 transcripts was monitored by real-time reverse transcriptase PCR of trophozoites using primers specific to the
glmbp1 gene (
Fig. 3A). The level of
tim transcript encoding triose 1-phosphate isomerase was used to normalize other transcripts from identical RNA samples [
14]. The crossing point-PCR cycle (Cp) values were estimated as 18.8 and 35.6 for
tim and
glmbp1, respectively. Thus, the level of
glmbp1 transcript expression was lower than that of
tim, but
glmbp1 transcripts were present in
G. lamblia trophozoites.
In addition, the presence of GlMBP1 in
Giardia trophozoites was demonstrated by western blot analysis of
Giardia extracts with anti-GlMBP1 antibodies. An immunoreactive protein of 48 kDa, the expected size of GlMBP1, was apparent (
Fig. 3B).
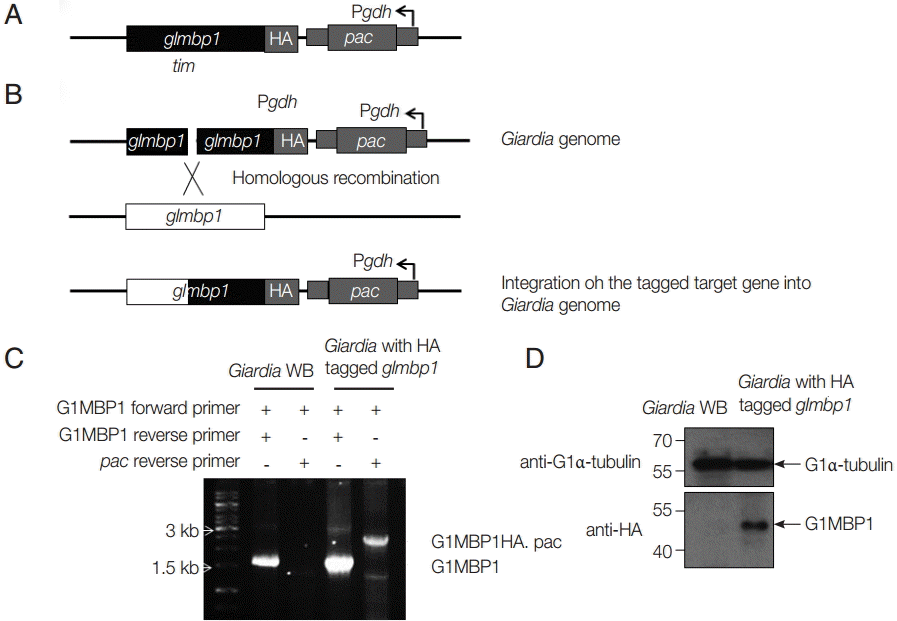
In the next experiment, we examined whether GlMBP1 was localized in the cytoskeletal structures of
G. lamblia. First, the
glmbp1 gene tagged with HA epitopes was integrated by electroporation of linear
glmbp1-HA DNA into the genomes of
G. lamblia trophozoites (
Fig. 4A,
B) [
15]. Integration of the chimeric
glmbp1-HA gene into the
Giardia chromosome was examined by PCR using
glmbp1-specific primers and a primer specific for the vector plasmid, the pac reverse primer (
Fig. 4C). A 1.6 kb
glmbp1 PCR product was detected in both
G. lamblia WB as well as in pGlMBP1HA.pac-transfected cells. A 2.5 kb PCR product, the expected length of
glmbp1 and the integrated plasmid, was amplified only from the genomic DNA of
G. lamblia transfected with pGlMBP1HA, but not from that of
G. lamblia WB.
Giardia trophozoites with the integrated HA-tagged
glmbp1 gene were then examined for the expression of a chimeric GlMBP1-HA by western blotting with anti-HA antibodies (
Fig. 4D). An immunoreactive protein band of 48 kDa was only present in the extracts of
Giardia with the integrated HA-tagged
glmbp1 gene (
Fig. 4D).
Giardia WB extracts did not show the immunoreactive protein. These results indicated that the HA-tagged
glmbp1 and pac genes were integrated into a
G. lamblia chromosome, and that HA-tagged GlMBP1 was expressed in
G. lamblia transfected with linear pGlMBP1HA.pac.
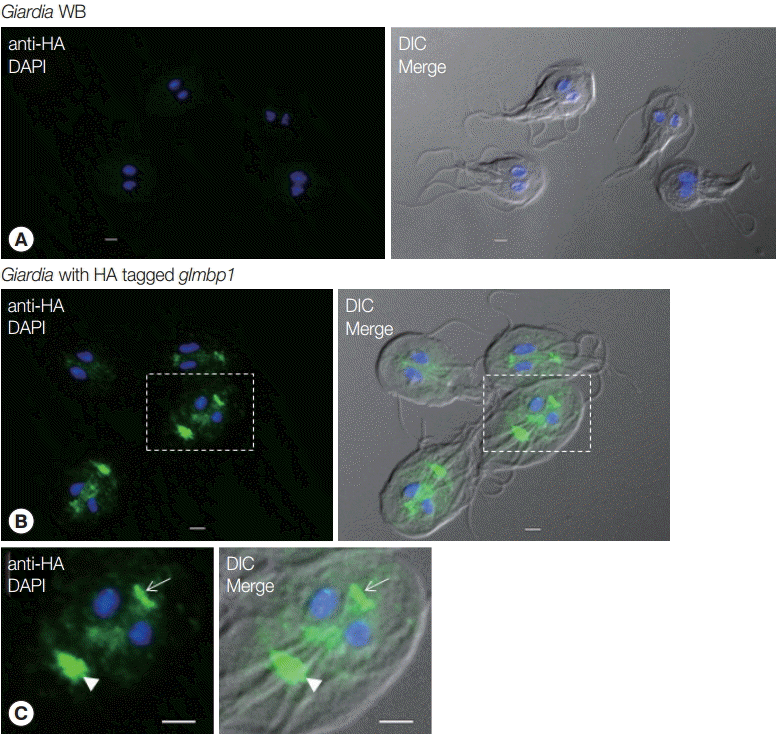
IFA was performed on
G. lamblia WB and
G. lamblia with the integrated
glmbp1-HA gene using anti-HA antibodies (
Fig. 5).
G. lamblia WB trophozoites did not show any fluorescence in the IFA (
Fig. 5A). In contrast, transgenic
G. lamblia with HA-tagged
glmbp1 showed green fluorescence at axonemes, median bodies, and basal bodies (
Fig. 5B,
C). This result indicated that GlMBP1 was located in cytoskeletal structures of
G. lamblia trophozoites.
DISCUSSION
The cytoskeleton of
G. lamblia is largely made of MTs, which includes a ventral disc, a median body, 8 basal bodies, and 4 pairs of flagella (anterior, posterior-lateral, ventral, and caudal). Each of the flagella is extended from an axoneme that is templated in a basal body. Basal bodies are conserved MT-organizing centers that nucleate structural apparatus such as flagella and cilia, and that function as spindle poles during cell division [
16]. In
G. lamblia, these structures were shown to be associated with MTs by electron immunocytochemistry [
17] as well as IFA using anti-tubulin antibodies (Kim and Park, an unpublished result). Interestingly, γ-tubulin was found by IFA using monoclonal antibodies specific to γ-tubulin, only in the basal bodies of ventral and posterior-lateral flagella in the interphase
Giardia cells. Further, this protein disappeared at early mitosis and reappeared in late-mitotic
G. lamblia [
18]. An extensive analysis of
G. lamblia, based on
in silico and proteomic methods, revealed 75 homologs of conserved basal body proteins. Thirteen of these homologs were found, by confocal microscopy, to co-localize with centrin in
Giardia basal bodies [
19]. Even though an association between GlMBP1 and basal bodies has not been reported, one of the MT-associated proteins was found to localize to cytoskeletal structures (axonemes, basal bodies, and median bodies) (
Figs. 2,
5B).
In vitro interaction of GlMBP1 with MTs (
Fig. 2) and its localization in the
Giardia cytoskeleton indicates a role for GlMBP1 in cell cycle-related functions. However, the information available on GlMBP1 is too limited to allow a conclusion about its relationship with
G. lamblia cell cycle. Monitoring the intracellular locations of GlMBP1 at various stages of
G. lamblia will be essential to defining its involvement in cell cycle control in this organism. In addition, knockdown of
glmbp1 expression will provide insight into the function of this protein in
G. lamblia.
In summary, a novel binding protein was isolated while screening for MT-binding protein. Association of this protein with MTs was confirmed by in vitro MT-binding assays using rGlMBP1 and polymerized MTs. The intracellular location of this protein in G. lamblia was observed using transgenic G. lamblia in which an HA-tagged glmbp1 gene was integrated into a chromosome. IFAs using anti-HA antibodies revealed that GlMBP1 is located in cytoskeletal structures, median bodies, basal bodies, and axonemes. The physiological role of this GlMBP1-MT interaction should be defined in future investigations.
Notes
-
We have no conflict of interest related to this work.
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1D1A1A09057595).
Fig. 1.Identification of GlMBP1, an MT-binding protein from Giardia extracts via in vitro MT-binding assays. (A) Trophozoites of G. lamblia WB were resuspended in PBS, and lysed by sonication. The binding of Giardia lysates to polymerized MTs was assessed in vitro using a Microtubule-Binding Protein Spin-Down Assay Kit BK029 (Cytoskeleton). Twenty µmoles of MTs was incubated with 100 µg of Giardia lysates in a total volume of 50 µl at room temperature for 40 min. The reaction mixtures were then centrifuged through a 50% glycerol cushion-PEM-taxol mixture at 100,000 g at 25˚C for 40 min using an ultracentrifuge. The pellet fraction was then resolved on 8% SDS-PAGE and visualized by silver staining. A sample with the same amount of Giardia extracts was precipitated by ultracentrifugation and compared side-by-side with the extracts precipitated with MTs. (B) An extended view of the SDS-PAGE gel showing proteins between 55 kDa and 25 kDa. (C) The protein band present only in the MT fraction was excised and digested with trypsin. The trypsin-treated proteins were analyzed by quadrupole time-of-flight (Q-TOF), in addition to matrix-assisted laser desorption ionization-TOF mass spectrometry (MALDI-TOF MS). For the Q-TOF liquid chromatography-tandem MS (LC-MS/MS) data sets, tandem mass spectra were submitted to our MASCOT in-house database search engine (NCBI NR database downloaded on 31 July 2009). For protein identification, a MASCOT ion score of >37 was used as the criterion for a meaningful result.

Fig. 2.In vitro MT-binding assays using rGlMBP1. Ten µg of rGlMBP1 was incubated without or with taxol-stabilized bovine MTs (20 µM), divided into pellet (P) and soluble (S) fractions by ultracentrifugation, and then separated by 12% SDS-PAGE. (A) A SDS-PAGE gel stained with Coomassie brilliant blue. (B) Western blot using anti-histidine antibodies (1:5,000 dilution). An arrowhead (about 55 kDa) indicates MTs, whereas arrows denote rGlMTBP.

Fig. 3.In vivo expression of GlMBP1 in
G. lamblia trophozoites. (A) Quantitative measurement of GlMBP1 transcripts. Total RNA was isolated from
G. lamblia using TRIzol. cDNA was synthesized from 5 µg of RNA using the ImProm-II
TM RT system and then analyzed with the Light Cycler 480 II Real-Time PCR System using LightCycler 490 DNA SYBR Green I Master (Roche Applied Science). Conditions for real-time PCR were as follows: pre-incubation at 95˚C for 5 min followed by 45 amplification cycles of 95˚C for 10 sec, 56˚C for 20 sec, and 72˚C for 10 sec. Real-time PCR was carried out in triplicate in a 96-well plate using the specific primers listed in
Table 1. The tim gene encoding triose-1-phosphate isomerase of
G. lamblia was used as an endogenous control for the reactions. (B) Western blot analysis. Ten µg of
Giardia extracts was separated by 12% SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was incubated with anti-GlMBP1 antibodies (1:1,000 dilution), followed by secondary antibodies (1:1,000 dilution).

Fig. 4.Expression of HA-tagged GlMBP1 in G. lamblia. (A) A schematic diagram of the plasmid pGlMBP1HA.pac. This plasmid contains the in-frame N-terminal end of the glmbp1 gene with 3 HA epitopes and the puromycin N-acetyltransferase gene (pac) cassette as a selective marker expressed from the glutamate dehydrogenase gene (gdh) promoter, Pgdh. (B) A strategy to integrate pGlMBP1HA.pac into a Giardia chromosome. Closed boxes show the plasmid sequences, and open boxes indicate Giardia chromosomal sequences. HA-tagging sequences and the pac cassette are indicated by gray boxes. (C) Genomic PCR analysis. PCR was performed using the GlMBP-F and GlMBP-HA-R or pac down primers on genomic DNA from G. lamblia WB cells or pGlMBP1HA.pac-transfected cells. (D) Western blot analysis. Ten µg of Giardia extracts (G. lamblia WB cells or pGlMBP1HA.pac-transfected cells) was separated by 12% SDS-PAGE and transferred to nitrocellulose membranes. The membrane was incubated with anti-HA antibodies (1:1,000 dilution), and then secondary antibodies (1:1,000 dilution).

Fig. 5.Localization of GlMBP1 in G. lamblia trophozoites. G. lamblia cells expressing HA-tagged GlMBP1 were fixed with chilled 100% methanol, and then permeabilized with PBS/0.5% Triton X-100. The cells were reacted with mouse anti-HA antibodies (1:50 dilution) and then incubated with AlexaFluor 488-conugated anti-mouse IgG (1:200). The cells were mounted on slides with VECTASHIELD Antifade Mounting Medium with 4ʹ,6-diamidino-2-phenylindole, and then observed with an Axiovert 200 fluorescence microscope (Carl Ziess). Differential interference contrast (DIC) images showed cell morphology. (A) IFA on G. lamblia WB using anti-HA antibodies. The bars indicate 2 µm. (B, C) IFA on G. lamblia with integrated HA-tagged glmbp1 using anti-HA antibodies. The bars indicate 2 µm. Arrowheads indicate median bodies, while arrows show basal bodies.

Table 1.Strains, primers, and plasmids used in this study
Table 1.
|
Strains/primers/plasmids |
Relevant characteristicsa
|
Source or reference |
|
G. lamblia
|
|
|
|
ATCC 30957 |
Clinical isolate |
ATCC |
|
E. coli
|
|
|
|
DH5a |
supE44 DlacU169 (F80 lacZ DM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1
|
Invitrogen |
|
BL21 (DE3) |
F’, ompT, hsdSB(rB-mB-) gal, dcm (DE3)
|
Invitrogen |
|
Primers |
|
|
|
8405F |
GCGAATTCGATGTCGATGGACGTTCCTA (EcoRI) |
|
|
8405R |
AGTTTAGCGGCCGCGCGCTTTGAGCCACACTCCA (NotI) |
|
|
GlMBP-F |
CGATCCATGGCCGGCTCTTGTCCAACTGCT (NcoI) |
|
|
GlMBP-HA-R |
GTTACGCGGCCGCTTAAGCGTAATCTGGAACATCG |
|
|
TATGGGTAAGCGTAATCTGGAACATCGTATGGGTAA |
|
|
GCGTAATCTGGAACATCGTATGGGTAGCGCTTTGAGCCACACTCCA (NcoI) |
|
|
pac down |
CGCGAATTCTCAGGCACCGGGCTT |
|
|
RT-tim_F |
CGAAAGTGGTTTGCGGAGAAG |
|
|
RT-tim_R |
CTATGTACGGGTCTTCGTAAGA |
|
|
RT-GlMBP-F |
GATGAAGTAGATAAGGCGGCA |
|
|
RT-GlMBP-R |
GAGCCACACTCCATACAGAAT |
|
|
Plasmids |
|
|
|
pET21b |
Expression vector for a histidine-tagged protein |
Novagen |
|
pETGlMBP1 |
pET21b, 1,338 bp encoding GlBMP1 (GiardiaDB; GL50803_8405) |
This study |
|
pGFP.pac |
Shuttle vector, AmpR, pac gene |
Singer et al. [13] |
|
pGlMBP1HA.pac |
pGFP.pac, 1,538 bp encoding GlMBP1 from its own promoter |
This study |
References
- 1. Elmendorf HG, Dawson SC, McCaffery JM. The cytoskeleton of Giardia lamblia. Int J Parasitol 2003;33:3-28.
- 2. Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 1997;13:83-117.
- 3. Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 2008;9:309-322.
- 4. Dawson SC, Sagolia MS, Mancuso JJ, Woessner DJ, House SA, Friz-Laylin L, Cande WZ. Kinesin-13 regulates flagellar, interphase, mitotic microtubule dynamics in Giardia intestinalis. Eukaryot Cell 2007;6:2354-2364.
- 5. Kim J, Nagami S, Lee KH, Park SJ. Characterization of microtubule-binding and dimerization activity of Giardia lamblia endbinding 1 protein. PLoS One 2014;9:e97850.
- 6. Kim J, Sim S, Kim J, Song K, Yong TS, Park SJ. Giardia lamblia EB1 is a functional homolog of yeast Bim1p that binds to microtubules. Parasitol Int 2008;57:465-471.
- 7. Holberton DV, Ward AP. Isolation of the cytoskeleton from Giardia. Tubulin and a low-molecular-weight protein associated with microribbon structures. J Cell Sci 1981;47:39-166.
- 8. Lourenço D, Andrade Ida S, Terra LL, Guimarães PR, Zingali RB, de Souza W. Proteomic analysis of the ventral disc of Giardia lamblia. BMC Res Notes 2012;5:41.
- 9. Hagen KD, Hirakawa MP, House SA, Schwartz CL, Pham JK, Cipriano MJ, De La Torre MJ, Sek AC, Du G, Forsythe BM, Dawson SC. Novel structural components of the ventral disc and lateral crest in Giardia intestinalis. PLoS Negl Trop Dis 2011;5:e1442.
- 10. Woessner DJ, Dawson SC. The Giardia median body protein is a ventral disc protein that is critical for maintaining a domed disc conformation during attachment. Eukaryot Cell 2012;11:292-301.
- 11. Keister DB. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg 1983;77:487-488.
- 12. Harlow E, Lane D. Immunizations. In Antibodies: A Laboratory Manual, Immunization. New York, USA. Cold Spring Harbor Laboratory; 1988, pp 53-63.
- 13. Singer SM, Yee J, Nash TE. Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol Biochem Parasitol 1998;92:59-69.
- 14. Kim J, Bae S, Sung M, Lee K, Park SJ. Comparative proteomic analysis of trophozoites versus cysts of Giardia lamblia. Parasitol Res 2009;104:475-479.
- 15. Gourguechon S, Cande WZ. Rapid tagging and integration of genes in Giardia intestinalis. Eukaryot Cell 2011;10:142-145.
- 16. Debec A, Sullivan W, Bettencourt-Diaz M. Centrioles: active players or passengers during mitosis? Cell Mol Life Sci 2010;67:21173-2194.
- 17. Crossley R, Marshall J, Cark JT, Holberton DV. Immunocytochemical differentiation of microtubules in the cytoskeleton of Giardia lamblia using monoclonal antibodies to γ-tubulin and polyclonal antibodies to associated low molecular weight proteins. J Cell Sci 1986;80:233-252.
- 18. Nohynkova E, Draber P, Reischig J, Kulda J. Localization of gamma-tubuin in interphase and mitotic cells of a unicellular eukaryote, Giardia intestinalis. Eur J Cell Biol 2000;79:438-445.
- 19. Lauwaet T, Smith AJ, Reiner DS, Romijn EP, Wong CCL, Davids BJ, Shah SA, Yates JR, Gillin FD. Mining the Giardia genome and proteome for conserved and unique basal body proteins. Int J Parasitol 2011;41:1079-1092.