Abstract
This is the first study to report Chrysomya pinguis (Walker) and Lucilia porphyrina (Walker) (Diptera: Calliphoridae) as forensically important blow fly species from human cadavers in Thailand, in addition to Chrysomya villeneuvi (Patton) already known in Thailand. In 2016, a fully decomposed body of an unknown adult male was discovered in a high mountainous forest during winter in Chiang Mai province. The remains were infested heavily with thousands of blow fly larvae feeding simultaneously on them. Morphological identification of adults reared from the larvae, and molecular analysis based on sequencing of 1,247 bp partial mitochondrial cytochrome c oxidase subunit 1 gene (CO1) of the larvae and puparia, confirmed the above mentioned 3 species. The approving forensic fly evidence by molecular approach was described for the first time in Thailand. Moreover, neighbor-joining phylogenetic analysis of the CO1 was performed to compare the relatedness of the species, thereby affirming the accuracy of identification. As species of entomofauna varies among cases in different geographic and climatic circumstances, C. pinguis and L. porphyrina were added to the list of Thai forensic entomology caseworks, including colonizers of human remains in open, high mountainous areas during winter. Further research should focus on these 3 species, for which no developmental data are currently available.
-
Key words: Chrysomya pinguis, Chrysomya villeneuvi, Lucilia porphyrina, forensic entomology, molecular identification, Thailand
INTRODUCTION
The contribution of forensic entomology has increased dramatically in many parts of the world. Flies of forensic importance are the subject of interest, since their specimens collected from human cadavers can be used in forensic investigations for estimating a minimum postmortem interval (PMI
min). As insect communities differ across seasons, the records of entomofauna collected from various death scenes in Thailand add to datasets that can be applied to countries with similar geography or climate. Fly larvae collected from actual forensic cases provide excellent opportunities. In Thailand, most case scenes are in urban, suburban, and forested areas, and have been based on forensic entomology casework reports since 2000 [
1]. Conversely, cases in high mountainous habitats are rare. Therefore, this study reports a case in a high mountainous area during winter, with the aim of adding information pertaining to species of forensic importance. This is a case of blow flies (Diptera: Calliphoridae),
Chrysomya pinguis (Walker),
Chrysomya villeneuvi (Patton), and
Lucilia porphyrina (Walker), being found on human remains, 2 of which have not been reported in Thailand before.
CASE RECORD
The corpse of an unknown male was discovered on the ground in a high open mountainous area (−1,200 m asl.) in Chiang Mai province, northern Thailand (
Fig. 1), during the winter of 2016. The temperature at the death scene ranged from 15 to 18°C. During the autopsy at the Department of Forensic Medicine, Chiang Mai University, the remains appeared as advanced stage of decomposition, with most of the body blackened. The skin and tissue at the neck and thorax were missing, thus the thoracic vertebrae and ribs were visible. Entomological specimens were collected during the autopsy, and remarkably, thousands of blow fly larvae (Diptera: Calliphoridae) were found feeding heavily on and/or scattered along the body. All stages of larvae (first, second, and third instars) and puparia coexisted on the corpse.
Some of the third instar larvae and puparia were collected randomly from the corpse and transported to the Department of Parasitology, Faculty of Medicine, Chiang Mai University. The obtained puparia were grouped into hairy pupae and non-hairy pupae. Both groups were reared separately in the transparent box laid with sawdust, until their adulthood was morphologically identified. In case of non-emerging adults, whole pupae were processed using molecular approaches as described below in order to prove the exact species.
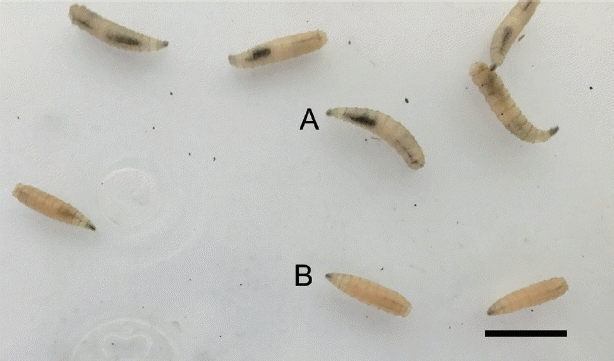
For third instar larvae, they were morphologically divided into 3 groups, hairy larvae (
Fig. 2, upper), creamy white non-hairy larvae (
Fig. 3A), and pinkish white non-hairy larvae (
Fig. 3B). Some larvae of each group were separately reared in the laboratory until adulthood for species verification. Meanwhile, the rest of them were dissected for morphological identification. Briefly, after placing some larvae in near-boiling water, the dead larvae were then cut across the middle of the second thoracic segment (TS2) and 7th abdominal segment (AS7), using a sharp surgical blade at the 2 sites, in order to view the anterior and posterior spiracles, respectively. Incomplete peritreme, a unique characteristic of maggots in the subfamily Chrysomyinae, was found at the posterior spiracle of the hairy larvae, revealing the species as
C. villeneuvi [
2]. However, the creamy white and the pinkish white of non-hairy larvae contrarily possessed incomplete and complete peritremes, respectively, defining as 2 distinct unidentified species. Apart from the TS2 and AS7 segments, the residual parts of each dissected larva were preserved in 70% ethanol and subsequently used to precisely confirm the species by molecular techniques.
Some larvae or pupae that were kept alive were successfully reared into mature adults and were morphologically approved for their species using taxonomic keys [
3], revealing the species as
C. villeneuvi and
C. pinguis. Briefly, adults of both species were metallic in coloration, presenting setulae on the posterodorsal surface of stem-vein of wing, prealar knob with erect hairs, and poorly developed bristles on the thoracic dorsum. Both male and female
C. villeneuvi had dichoptic eyes, fuscous gena, and brown to black mesothoracic spiracles. Femora were greatly swollen in both sexes, but more noticeable in males. The disc of the 5th tergite of males was covered with dense short hairs, while almost bare in females. As for
C. pinguis, males had holoptic eyes, while dichoptic in females. Gena was fuscous (brownish gray to black). Anterior half of the upper squama was covered with black hairs. Posthumeral bristles were well developed. The disc of the 5th tergite was covered with many fine erect bristles in both sexes.
To confirm the species level, genomic DNA was extracted from the immature stages (6 third instars and 18 pupae) using the E.Z.N.A.® Tissue DNA Kit (Omega Biotek, Germany), according to the manufacturer’s protocols. The extracted DNA was eluted in 100 μl elution buffer of which 5 μl further used for PCR amplification.
Partial sequences of the mitochondrial cytochrome
c oxidase subunit 1 gene (
CO1) were amplified using a forward and a reverse primer (TY-J-1460 and C1-N-2800, respectively) formerly designed for calliphorid identification [
4]. Reaction mixtures of the PCR were prepared on a 50 μl scale containing 5 μl of template DNA, 1× PCR buffer, 0.2 mM dNTPs, 1.5 mM MgCl
2, 0.4 μM of each primer, and 1 unit of Platinum
® Taq DNA polymerase (Invitrogen, Gaithersburg, Maryland, USA). The thermal cycler condition consisted of a pre-denaturation step at 95°C for 5 min, followed by 35 cycles at 95°C for 1 min, 49°C for 2 min, 72°C for 2 min, and final extended step at 72°C for 7 min, and was implemented in a TPersonal Combi Thermo Cycler (Biometra, Göttingen, Germany). The amplified PCR products were electrophoretically run in 1% agarose gel containing Redsafe
® Nucleic Acid Staining Solution (iNtRON Biotechnology, Seoul, Korea), and visualized under UV illumination for DNA fragment detection.
The PCR products were purified by the E.Z.N.A.® Cycle Pure Kit (Omega Biotek), according to the manufacturer’s instructions, and sent on to First BASE Laboratories Sdn Bhd (Selangor, Malaysia) for DNA sequencing, principally performed using BigDye® Terminator v3.1 cycle sequencing kit chemistry. Purified PCR products were sequenced in both directions using the identical pair of primers utilized in the PCR.
Consensus of partial
CO1 sequences were achieved by assembling data of forward and reverse sequences using SeqScape v2.5.0 (Applied Biosystems Foster City, California, USA) and BioEdit v7.0.9.0 software [
5]. The sequences were submitted to the GenBank database under the accession nos. KX096338 -KX096346. Searches for the highest similarity between queried Thai blow fly sequences, and available biological sequences in the GenBank database, were performed separately, using a BLAST search at the National Center for Biotechnology Information (
http://blast.ncbi.nlm.nih.gov/Blast.cgi).
The PCR products were retrieved successfully from all 6 larvae and 4 out of 18 pupae. Almost full length of
CO1 gene (1,247 bp) were sequenced from purified PCR products, excluding 1 pupa (P8), of which only 1,213 bp in length was achieved. The consequences of successful sequence analysis are summarized in
Table 1. Based on a BLAST search, 2 genera comprised 3 species of
C. pinguis,
C. villeneuvi, and
L. porphyrina, were identified in this study. All analyzed
CO1 sequences showed the highest identity of up to 99–100%; with most of them top rank hitting
CO1 sequences of the Thai blow flies from Chiang Mai that were submitted previously to GenBank. Within the same species, the intraspecific divergence in
CO1 of
C. pinguis and
L. porphyrina were detected, with 9 and 20 nucleotide differences in positions of each species, respectively.
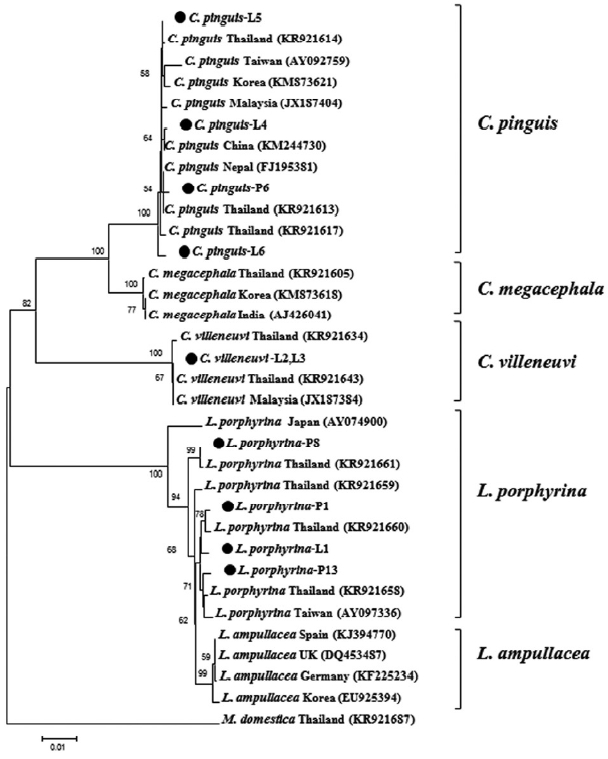
In addition, neighbor-joining (NJ) phylogenetic tree based on the
CO1 sequences (1,213 bp) of Thai blow fly species and available data from the GenBank, was constructed in the MEGA6 [
6], using the Kimura 2-parameter (K2P) model with 1,000 bootstrap replicates. The
CO1 sequences of
Chrysomya megacephala (Fabricius) and
Lucilia ampullacea (Villeneuve) which are closely related species to
C. pinguis and
L. porphyrina, respectively, were also added into the analysis, together with an outgroup sequence of a house fly,
Musca domestica (Linnaeus). According to the tree (
Fig. 4), each blow fly species, except
L. porphyrina and
L. ampullacea, could be clearly separated into its own monophyletic clade with highly bootstrap value (100%).
DISCUSSION
When comparing all forensic entomology cases analyzed in Thailand since 2000, this study was the first to detect
C. pinguis and
L. porphyrina in human remains under tropical conditions during winter in a high mountainous area. In past forensics,
C. megacephala and
Chrysomya rufifacies (Macquart) were the predominant species associated with human cadavers at variable ecological altitudes in both urban and high mountainous areas of Thailand [
1]. Entomological surveys of possible forensically important fly species in northern Thailand found that
C. pinguis tended to prevail in highland areas of mixed deciduous forests, which was similar to
Chrysomya thanomthini (Kurahashi & Tumrasvin),
Hypopygiopsis tumrasvini (Kurahashi),
Lucilia papuensis (Macquart), and
L. porphyrina [
7].
C. pinguis has been shown to colonize human corpses during crime scene investigations in documentation from Japan [
8], Malaysia [
9], and South Korea [
10]. In Taiwan, this blow fly species was important when used to estimate the PMI
min in the cool season [
11]. Much less is known about the bionomics of
C. pinguis.
The identification of blow flies in this study was based on the morphology of larvae sampled from the corpse and adults reared from immature stages, and affirmed by molecular analysis. It should be noted that during entomological sampling from a forensic autopsy, the third instar of
C. pinguis cannot be distinguished from a closely related species, such as
C. megacephala. Even when the main characteristics for identification can be observed under light microscopy, 9–11 papillae of the anterior spiracle of
C. pinguis were found, which was comparable with the 8–12 seen in
C. megacephala [
12]. Furthermore, great similarity of the spines between the first and second thoracic segments and morphology of the posterior spiracle makes it difficult to perform differential identification [
12]. Finally, rearing larvae to adults confirmed the species of
C. pinguis, which was verified later by molecular analysis. Moreover, molecular analysis was also helpful for species approving, particularly in unsuccessful rearing of some species, such as
L. porphyrina in this study.
Because the conspecific variation in
CO1 gene has been reported from the genus Chrysomya and
Lucilia [
13,
14], the phylogenetic relationship among 3 Thai blow fly species (
C. pinguis,
C. villeneuvi, and
L. porphyrina) together with the related species from other countries was then analyzed. The usefulness of
CO1 barcoding as a potential identification tool was shown convincingly, splitting each fly genus into separated clade. However,
CO1 was not completely distinguishable the species of
L. porphyrina from
L. ampullacae and sorted them into the same
Lucilia clade as closely related sister species (
Fig. 4). For
L. porphyrina, a high intraspecific variation within Thai isolates has recently been documented [
15]. Hence, it was not surprisingly that the differences up to 20 nucleotide positions in
CO1 sequences among Thai
L. porphyrina were observed in this study. Furthermore, the BLAST results showed non-identical (100%)
CO1 sequence from GenBank, but only 99% identity of other Thai
L. porphyrina isolates was hit (
Table 1). To our knowledge,
L. ampullacae had never been reported from Thailand. However, the species was known to be the forensically important blowfly indicator of the rural sites in Germany [
16] and Western Europe, being abundantly in summer, especially at low altitudes (100 and 500 m asl.) [
17]. Reasonably for the present study, the molecular based identification of
L. porphyrina may be efficient enough for species validation, particularly in Thailand where
L. ampullacae does not coexist. To discriminate closely related sister species in co-exiting area, phylogenies based on more than 1 genetic marker merits investigation to retrieve sufficient genetic information in different loci of genes [
18].
Despite flies of forensic importance being known, this study was the first to confirm the presence of
L. porphyrina in human remains in Thailand, especially by molecular approach, thereby listing the forensic importance of this species in the country. Its collection at the death scene in this case is comparable to that in several studies of colonized human cadavers and/or carcasses. In Nagasaki prefecture, western Japan, the upper region of mountainous areas was considered a primary habitat of this blow fly species [
19]. In Taiwan, this species was observed only in the northern region during winter [
11]. It also was collected for the list composed of sarcosaprophagous fly species in the community by using animal carcasses as bait in Guiyan in China [
20], Pakistan [
21], and Malaysia [
22]. This species was recorded to inhabit primarily forest areas, and adults are attracted to decaying animal matter, while larvae are scavengers [
23].
Among seasons, blow fly collections during the winter were low when compared with summer and rainy seasons [
24,
25]. It is interesting that
C. villeneuvi,
C. pinguis, and
L. porphyrina coexisted in large numbers during the winter in high mountainous areas of northern Thailand. Not surprisingly, outdoor human remains are infested by huge numbers of various common corpse-visiting fly species [
1,
9] of which up to 4 species were found to coexist within a single human corpse [
1].
Investigation in this study indicated a higher abundance of non-hairy maggots, when compared to the hairy-maggot of C. villeneuvi in the current casework. However, this study observed a small number of C. villeneuvi puparia from the remains, thus assuming potential behavior as a primary invader. However, due to the lack of developmental information on these 3 species (C. villeneuvi, C. pinguis, and L. porphyrina) in Thailand, estimating the PMImin in this case was problematic for forensic investigations. Therefore, correct identification of fly larvae collected from human remains, together with developmental datasets of the 3 species, would be needed for applying in forensic investigations, particularly when estimating the PMImin.
Notes
-
CONFLICT OF INTEREST
We have no conflict of interest related to this work.
ACKNOWLEDGMENT
The authors are grateful to the Faculty of Medicine and Chiang Mai University, Thailand for supporting and defraying the publication cost.
Fig. 1Map of mountainous area (~1,200 m asl.) of Chiang Mai province, northern Thailand, where the human remains were discovered.
Fig. 2Hairy third instar (upper) and puparium (lower) of Chrysomya villeneuvi collected from the human remains.

Fig. 3Non-hairy third instar of unidentified larvae collected from the corpse showing 2 groups of yellowish-white larvae (upper A) and pinkish-white larvae (lower B). Scale bar=1 cm.

Fig. 4Neighbor-joining tree based on the CO1 barcoding sequences (1,213 bp) showing phylogenetic relationships among Thai blow flies identified from the corpse and related species from other countries. The bootstrap values higher than 50% are shown above the node. Bar displays 0.01 substitutions per site.

Table 1Species identification of immature stages of Thai blow flies collected from human remains based on partial COI sequences (1,247 bp)
a
Table 1
|
Stages of fly |
Code no. of specimens |
Submitted accession no. |
Species identification |
Accession no. of GenBank Reference (% identity)b
|
|
Larva |
L1 |
KX096338 |
L. porphyrina
|
KR921659 (99) |
|
L2 |
KX096339 |
C. villeneuvi
|
KR921643 (100) |
|
L3 |
KX096339 |
C. villeneuvi
|
KR921643 (100) |
|
L4 |
KX096340 |
C. pinguis
|
KM244730 (99) |
|
L5 |
KX096341 |
C. pinguis
|
KR921614 (99) |
|
L6 |
KX096342 |
C. pinguis
|
KR921617 (99) |
|
|
Pupa |
P1 |
KX096343 |
L. porphyrina
|
KR921660 (99) |
|
P6 |
KX096344 |
C. pinguis
|
KR921613 (99) |
|
P8 |
KX096346 |
L. porphyrina
|
KR921661 (99) |
|
P13 |
KX096345 |
L. porphyrina
|
KR921658 (99) |
References
- 1. Sukontason K, Narongchai P, Kanchai C, Vichairat K, Sribanditmongkol P, Bhoopat T, Kurahashi H, Chockjamsai M, Piangjai S, Bunchu N, Vongvivach S, Samai W, Chaiwong T, Methanitikorn R, Ngern-Klun R, Sripakdee D, Boonsriwong W, Siriwattanarungsee S, Srimuangwong C, Hanterdsith B, Chaiwan K, Srisuwan C, Upakut S, Moopayak K, Vogtsberger RC, Olson JK, Sukontason KL. Forensic entomology cases in Thailand: a review of cases from 2000 to 2006. Parasitol Res 2007;101:1417-1423.
- 2. Sukontason K, Sukontason KL, Piangjai S, Narongchai P, Samai W, Boonchu N, Sripakdee D, Ngern-Klun R, Siriwattanarungsee S. Morphology of second and third instars of Chrysomya villeneuvi Patton (Diptera: Calliphoridae), a fly species of forensic importance. Forensic Sci Int 2005;154:195-199.
- 3. Kurahashi H, Boonchu N. The blow flies recorded from Thailand, with the description of a new species of Isomyia Walker (Diptera: Calliphoridae). Jpn J syst Ent 2011;17:237-278.
- 4. Sperling FA, Anderson GS, Hickey DA. A DNA-based approach to the identification of insect species used for postmortem interval estimation. J Forensic Sci 1994;39:418-427.
- 5. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999;41:95-98.
- 6. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013;30:2725-2729.
- 7. Moophayak K, Klong-Klaew T, Sukontason K, Kurahashi H, Tomberlin JK, Sukontason KL. Species composition of carrion blow flies in northern Thailand: altitude appraisal. Rev Inst Med Trop Sao Paulo 2014;56:179-182.
- 8. Saigusa K, Takamiya M, Aoki Y. Species identification of the forensically important flies in Iwate prefecture, Japan based on mitochondrial cytochrome oxidase gene subunit 1 (CO1) sequences. Leg Med (Tokyo) 2005;7:175-178.
- 9. Kavitha R, Nazni WA, Tan TC, Lee HL, Azirun MS. Review of forensically important entomological specimens collected from human cadavers in Malaysia (2005–2010). J Forensic Leg Med 2013;20:480-482.
- 10. Shin SE, Jang MS, Park JH, Park SH. A forensic entomology case estimating the minimum postmortem interval using the distribution of fly pupae in fallow ground and maggots with freezing injury. Korean J Leg Med 2015;39:17-21.
- 11. Lin IL YC, Pai CY, Shiao SF. Population analysis of forensically important Calliphoridae on pig corpses in Taiwan. J Forensic Sci 2010;9:25-34.
- 12. Sukontason K, Sukontason KL, Ngern-Klun R, Sripakdee D, Piangjai S. Differentiation of the third instar of forensically important fly species in Thailand. Ann Entomol Soc Am 2004;97:1069-1075.
- 13. Stevens JR, Wall R, Wells JD. Paraphyly in Hawaiian hybrid blowfly populations and the evolutionary history of anthropophilic species. Insect Mol Biol 2002;11:141-148.
- 14. McDonagh LM, Stevens JR. The molecular systematics of blowflies and screwworm flies (Diptera: Calliphoridae) using 28S rRNA, COX1 and EF-1α: insights into the evolution of dipteran parasitism. Parasitology 2011;138:1760-1777.
- 15. Zajac BK, Sontigun N, Wannasan A, Verhoff MA, Sukontason K, Amendt J, Zehner R. Application of DNA barcoding for identifying forensically relevant Diptera from northern Thailand. Parasitol Res 2016;115:2307-2320.
- 16. Fremdt H, Amendt J. Species composition of forensically important blow flies (Diptera: Calliphoridae) and flesh flies (Diptera: Sarcophagidae) through space and time. Forensic Sci Int 2014;236:1-9.
- 17. Zabala J, Díaz B, Saloña-Bordas MI. Seasonal Blowfly Distribution and Abundance in Fragmented Landscapes. Is It Useful in Forensic Inference about Where a Corpse Has Been Decaying? PLoS One 2014;9:e99668.
- 18. Williams KA, Lamb J, Villet MH. Phylogenetic radiation of the greenbottle flies (Diptera, Calliphoridae, Luciliinae). Zookeys 2016;568:59-86.
- 19. Suenaga O, Kurahashi H. Life cycle of an Oriental blow fly, Lucilia porphyrina (Walker) in Nagasaki, Western Japan. Trop Med 1995;37:99-107.
- 20. Chen L. The observation of sarcosaprophagous flies community composition, seasonal variation and growth of the length in the suburbs of Guiyang. Chin J Forensic Med 2011;26:204-206.
- 21. Shah ZA, Fatima Z. Impact of flesh age, trap color and decomposition stage on the population dynamics and species composition of calliphoridae and sarcophagidae. Pakistan J Zool 2007;39:45-50.
- 22. Silahuddin SA, Latif B, Kurahashi H, Heo CC. The Importance of habitat in the ecology of decomposition on rabbit carcasses in Malaysia: implications in forensic entomology. J Med Entomol 2015;52:9-23.
- 23. Kurahashi H, Chowanadisai L. Blow flies (Insecta: Diptera: Calliphoridae) from Indochina. Species Divers 2001;6:185-242.
- 24. Ngoen-klan R, Moophayak K, Klong-klaew T, Irvine KN, Sukontason KL, Prangkio C, Somboon P, Sukontason K. Do climatic and physical factors affect populations of the blow fly Chrysomya megacephala and house fly Musca domestica? Parasitol Res 2011;109:1279-1292.
- 25. Klong-Klaew T, Sukontason K, Ngoen-klan R, Moophayak K, Irvine KN, Kurahashi H, Prangkio C, Sanit S, Sukontason KL. Impact of abiotic factor changes in blowfly, Achoetandrus rufifacies (Diptera: Calliphoridae), in northern Thailand. Parasitol Res 2014;113:1353-1360.