Abstract
In this study, we describe Korean isolates of Trichomonas vaginalis infected with double-stranded (ds) RNA virus (TVV). One T. vaginalis isolate infected with TVV IH-2 evidenced weak pathogenicity in the mouse assay coupled with the persistent presence of a dsRNA, thereby indicating a hypovirulence effect of dsRNA in T. vaginalis. Cloning and sequence analysis results revealed that the genomic dsRNA of TVV IH-2 was 4,647 bp in length and evidenced a sequence identity of 80% with the previously-described TVV 1-1 and 1-5, but only a 42% identity with TVV 2-1 and 3 isolates. It harbored 2 overlapping open reading frames of the putative capsid protein and dsRNA-dependent RNA polymerase (RdRp). As previously observed in the TVV isolates 1-1 and 1-5, a conserved ribosomal slippage heptamer (CCUUUUU) and its surrounding sequence context within the consensus 14-nt overlap implied the gene expression of a capsid protein-RdRp fusion protein, occurring as the result of a potential ribosomal frameshift event. The phylogenetic analysis of RdRp showed that the Korean TVV IH-2 isolate formed a compact group with TVV 1-1 and 1-5 isolates, which was divergent from TVV 2-1, 3 and other viral isolates classified as members of the Giardiavirus genus.
-
Key words: Trichomonas vaginalis virus, TVV, dsRNA-dependent RNA polymerase, Totiviridae, ribosomal frameshift
INTRODUCTION
Trichomonas vaginalis is one of the principal worldwide human sexually transmitted disease agents (
WHO, 2001). Some isolates of this protozoan parasite have been shown to be infected by a double-stranded RNA (dsRNA) virus referred to as TVV (
Wang and Wang, 1985,
1986;
Wang et al., 1987). TVV was primarily described as a uniform population of icosahedral particles, 33 nm in diameter (
Wang and Wang, 1986). However, electron microscopic studies have recently shown that several types of virus-like particles (VLPs) could simultaneously infect
T. vaginalis isolates (
Bessarab et al., 2000;
Benchimol et al., 2002a,
2002b,
2002c). Although the most abundant type of virus detected was the aforementioned icosahedral particles, the number of dsRNA segments varied from 1 to 3 in different TVV isolates. Also, the length of those dsRNA segments varied from 4.3 to 5.0 kb (
Khoshnan and Alderete, 1993;
Su and Tai, 1996). This genomic complexity of TVV was confirmed by the results that the capsid proteins among the TVV isolates varied in size (75-85 kDa), as did their immunoreactions against capsid protein antibody (
Alderete et al., 2003).
Four species of genomic dsRNAs have been cloned from the TVV isolates 1-1, 1-5, 2-1 and 3, and their genetic information has been previously reported (
Tai
and Ip, 1995;
Su and Tai, 1996;
Bessarab et al, 2000) (GenBank Accession no. U08999, U57898, AF127178, and AF325840). The lengths of the dsRNAs varied in a range of 4,647 to 4,844 bp, but they shared the same gene organization of overlapping putative capsid protein and dsRNA-dependent RNA polymerase (RdRp). The capsid protein genes encode for 75-85 kDa proteins, and the RdRp genes encode for approximately 160 kDa of capsid protein-RdRp fusion protein, via a ribosomal frameshifting mechanism (
Dinman et al., 1991). The genomic organization of these isolates identified TVV as a legitimate member of the
Totiviridae family. Despite its rather simple viral gene organization, the role of the
T. vaginalis virus and its dsRNA in the gene expression of
T. vaginalis remain to be clearly delineated.
In the present study, we describe, for the first time, the presence of a virus in a Korean T. vaginalis isolate [designated TVV INHA(IH)-2], and verify its identity via comparisons of its properties with those of previously reported TVV isolates. Korean T. vaginalis isolates, with or without virus, have been assessed for pathogenicity in mice, and the complete cDNA sequence of the TVV IH-2 genomic dsRNA has been determined. The gene organization has been evaluated, and a phylogenetic analysis was conducted in order to compare its relationship with other TVV isolates, as well as with other members of the Totiviridae family.
MATERIALS AND METHODS
Culture of Korean T. vaginalis isolates
Korean
T. vaginalis isolates were collected from outpatients at the Inha University Hospital for standard clinical laboratory evaluations. Initially, the isolates were cultivated in trypticase-yeast extract-maltose (TYM) medium (
Diamond, 1957) supplemented with 10% heat-inactivated horse serum, gentamycin (10 µg/ml), and penicillin G (60 µg/ml), and then maintained in identical medium without antibiotics after axenization.
All Korean
T. vaginalis cultures were rapidly screened for TVV dsRNA as described by Wang et al. (
1987). In brief, more than 10
7 organisms were harvested and vortexed in 0.1 M sodium acetate, pH 5.0, and 1% sodium dodecyl sulfate. The lysates were extracted with water-saturated phenol at 65℃, and the total RNA was precipitated with ethanol. The presence of TVV dsRNA was determined via 1% agarose gel electrophoresis. In order to verify the relationship between the virus and the dsRNA, the TVV virus was purified from the IH-2 isolate, as previously described by Khoshnan and Alderete (
1993), with minor modifications. Approximately 10
9 organisms were collected and resuspended in TNM buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl
2). After sonication, the cell debris was removed via low-speed centrifugation, and the crude virus was pelleted through a 20% sucrose cushion for 2 hr at 100,000 g. The pellet was then resuspended in TNM buffer, layered on 10 to 50% (w/v) linear sucrose density gradients, and centrifuged for 3 hr at 140,000 g in a
Beckman SW41Ti rotor. The virus bands were collected, and concentrated via ultracentrifugation. RNA was extracted from the viral preparation and evaluated as described above.
In order to confirm that the RNA was double-stranded, its sensitivity to DNase and ribonuclease was assessed in accordance with the procedures previously described by Kim and Bozarth (
1985). The viral nucleic acid was incubated for 30 min with 1 Unit of RQ1 DNase (Promega, Madison, Wisconsin, USA), and with 0.5 µg/ml of pancreatic ribonuclease (Sigma, St. Louise, Massachusetts, USA) in 2 × SSC (0.3 M sodium acetate, 0.03 M sodium citrate, pH 7.4), and 0.01 × SSC buffers at 37℃. The integrity of the viral RNA was assessed via 1% agarose gel electrophoresis.
The pathogenicity of
T. vaginalis isolates in connection with the presence of dsRNA was assessed in accordance with the protocols described by Honigberg et al. (
1966). At the stage of 30 generations in cultivation, 10
6 organisms per ml of the selected isolates with over 95% viability were subcutaneously injected into mice. After 6 days, the number of mice evidencing abscessed was determined, and the volume of each abscess was measured using dial calipers. Fresh isolates represent organisms grown axenically for less than 10 generation after isolation, and long term grown parasites were passaged for over 6 mo.
In order to analyze the genomic organization of the TVV IH-2 Korean isolate, approximately 4.6 kb of dsRNA was cloned and sequenced on the basis of the techniques described by Kim et al. (
2003). The cDNA was synthesized from denatured dsRNA primed with random hexamer or gene-specific primers, using the Universal RiboClone cDNA system (Promega). Then the cDNA was ligated into pBlueScript II vector (Promega) and transformed into
Escherichia coli DH5α cells. The 5'-terminal sequences of dsRNA were cloned via 5'-RACE PCR (Rapid Amplification of cDNA Ends by Polymerase Chain Reaction) (
Frohman et al., 1988). All the PCR-amplified products were ligated into pGEM-T Easy II vector (Promega) and transformed into
E. coli DH5α cells. The cDNA clones were sequenced with an automated DNA sequencer (ABI, Foster City, California, USA). The putative proteins were then predicted using the ORF finder program, and compared with database information using the BLAST functions of the NCBI Web server (
http://www.ncbi.nlm.nih.gov).
RESULTS
T. vaginalis Korean isolates infected by dsRNA
A total of 152 samples were evaluated in 2004, and 22 were positive for
T. vaginalis. After the direct extraction of dsRNA from the 22
T. vaginalis isolates, only 3 (IH-1, IH-2, and IH-3) evidenced the presence of a 4.6 kb dsRNA segment (
Table 1). After several mo of serial in vitro cultivations, only 1 isolate (IH-2) maintained the dsRNA. The 4.6 kb dsRNA segment purified from the TVV preparation was also observed after electrophoresis in 1% agarose (
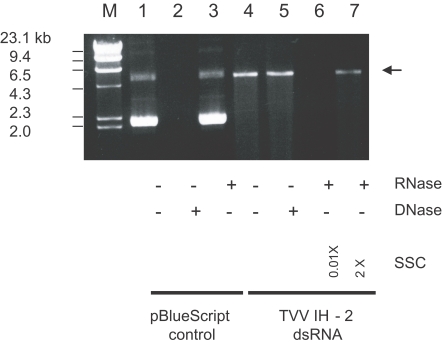
Fig. 1, lane 4). It co-migrated with the dsRNA extracted directly from the cell via the rapid screening technique. This result confirmed that the 4.6 kb dsRNA was a genomic dsRNA of TVV.
The stability of TVV nucleic acid to DNase indicated that this viral nucleic acid was RNA (
Fig. 1, lane 5). The stability in the presence of ribonuclease in high ionic strength buffer (2 × SSC) and its susceptibility in low ionic strength buffer (0.01 × SSC) verified that this viral RNA was double-stranded RNA (
Fig. 1, lanes 6 and 7).
Seven isolates, including 3 isolates with TVV dsRNA, were selected and assessed for pathogenicity in mice (
Table 1). The isolates were classified into 3 groups on the basis of the criteria established by Honigberg et al. (
1966). IH-3, IH-10 and IH-11 were grouped into the severe-pathogenic strains, IH-1, IH-6 and IH-8 were grouped into the moderate-pathogenic strains, and IH-2 was classified as a non/weak-pathogenic strain group. TVV dsRNA was observed in fresh IH-1, IH-2 and IH-3 isolates, but only IH-2 maintained dsRNA upon long term in vitro cultivation.
A full-length cDNA clone, pTVV IH-2, was obtained, and the size of the inserted cDNA was determined to be 4,647 bp (GenBank accession no. DQ270032). The conserved sequence motifs of 5'- ACTCAACATT-3' and 5'-CTATACCTTC-3' of TVV 1-1 and 1-5 isolates were also observed at the 5' and 3' termini of TVV IH-2, respectively. The identity percentage of the genomic dsRNA of TVV IH-2 was 80%, when compared to TVV 1-1 and 1-5 (
Tai and Ip, 1995;
Su and Tai, 1996), but only 42%, when compared to the TVV 2-1 and 3 isolates (
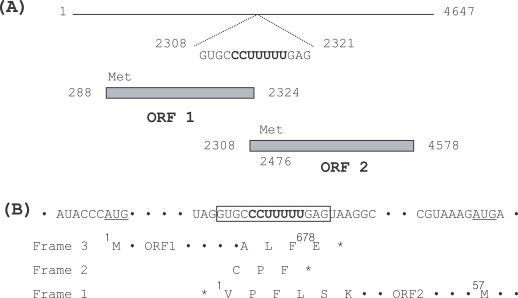
Bessarab et al., 2000). It harbored 2 overlapping open reading frames (ORF 1 and ORF 2) coupled with a ribosomal frame shift event on the plus strand viral RNA (
Fig. 2A). ORF 1 (nt 288-2,324) harbored 678 amino acid residues with a molecular weight of 74.6 kDa. Presumably, it could be translated into a capsid protein of the TVV IH-2 isolate, as the amino acid sequence shared a similarity of over 85% with the capsid proteins of the TVV 1-1 and 1-5 isolates. However, less than 17% similarity was observed when compared to the capsid proteins of the TVV 2-1 and 3 isolates. ORF 2 (nt 2,308-4,578), which overlapped ORF 1 in a +1 reading frameshift, commenced with a valine and terminated in a stop codon 70 nt from the 3'-terminus, but the first methionine codon (nt 2,476-2,478) was located at amino acid position 57 in this ORF. It harbored 756 amino acid residues, with a molecular weight of 85.7 kDa. A BLAST search of the deduced amino acid sequence of ORF 2 showed that it was a putative dsRNA-dependent RNA polymerase with a conserved RdRp motif (SG...T...DS...N and GDD) (
Poch et al., 1989), and it evidenced a similarity of 83% with the RdRp of the previously described TVV 1-1 and 1-5 isolates (
Tai and Ip, 1995;
Su and Tai, 1996). However, again only a 25% similarity was noted, when compared with the RdRp of TVV 2-1 and 3 isolates (
Bessarab et al., 2000). Within the conserved 14 nt overlap (nt 2,308-2,321) between the capsid protein and the RdRp genes, a potential ribosomal slippage heptamer (CCUUUUU) was identified (
Fig. 2B). The same heptamer and a very similar surrounding context have been previously reported in TVV1-1 and 1-5 isolates (
Tai and Ip, 1995;
Su and Tai, 1996). The adjacent termination codon (nt 2,305-2,307) in reading frame 1 was determined to precede the termination codon of ORF 1 in reading frame 3. Although it was not demonstrated that ribosomal frameshifting applied to the gene expression of the TVV IH-2 isolate, either 2 consecutive -1 shifts or a +1 shift event could be predicted, as was also the case with the TVV1-1, 1-5 and 2-1 isolates.
The genomic organization and genetic diversity of TVV resemble the non-segmented dsRNA viruses of the
Totiviridae family, including the
Saccharomyces cerevisiae virus (ScV),
Giardia lamblia virus (GLV) and
Leishmania RNA virus (LRV) (
Icho and Wickner, 1989;
Bruenn, 1993;
Wang et al., 1993;
Stuart et al., 1992;
Scheffter et al., 1994,
1995). The RdRps of ScV L-A, ScV L-BC, GLV, LRV 1-1, 1-4 and 2-1 were included in order to analyze the intergeneric relationship of these viruses in the
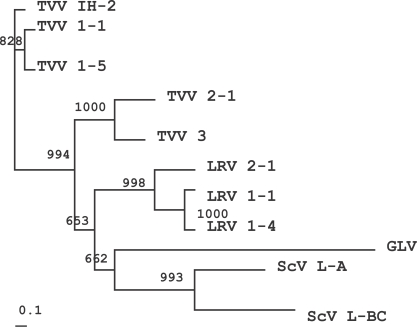
Totiviridae family (GenBank Accession no. J04692, U01060, NC 003555, NC 002063, NC 003601, NC 002064). The phylogenetic tree indicated that TVV IH-2 isolate was related closely to TVV 1-1 and 1-5, but not to TVV 2-1 and 3 (
Fig. 3). This tree was generally consistent with the sequence alignment results indicating an 83% similarity to TVV 1-1 and 1-5, and a 25% similarity to the TVV 2-1 and 3 isolates. Similar results were noted in previous reports, showing that the RdRp sequences of TVV 1-5 evidenced 84% identity with those of TVV 1-1, but TVV 2-1 shared an overall identity of 30% with TVV 1-1 (
Su and Tai, 1996;
Bessarab et al., 2000). This tree verified the previous results that the TVV 2-1 and 3 isolates were divergent from the TVV 1-1 and 1-5 isolates. Interestingly, however, the TVV 2-1 and 3 isolates were found to be accurately grouped with LRV, GLV, and ScV, as shown in the
Fig. 3. A similar phylogenetic tree was also observed upon comparisons of the capsid proteins (data not shown).
DISCUSSION
In this study, for the first time, the presence of the
T. vaginalis virus was identified in 3 Korean isolates. In particular, the IH-2 isolate maintained TVV upon long-term in vitro cultivation. Although it should be noted that our study was limited by its small sample size, the IH-2 isolate evidenced weak pathogenicity in mice. This hypovirulence effect was considered to be associated with the persistent presence of TVV or its genomic dsRNA, as published by the previous reports (
Alderete et al., 1986;
Wang et al., 1987;
Alderete and Neale, 1989;
Khoshinan and Alderete, 1994). In these reports they addressed the fact that the presence of TVV viruses or their genomic dsRNA directly influenced phenotypic variation via the regulation of the surface immunogen, P270, of the parasite. Also these authors defined 2 types of isolates on the basis of their effects on the synthesis of the surface immunogen type I isolates, which were demonstrated to harbor no viruses, were homogeneous populations of parasites that synthesized P270, but did not express this immunogen on the surface of the cell. However, the type II isolates comprised a heterogeneous population of parasites, some of which evidenced P270 on the surfaces. Only the type II isolates were determined to harbor TVV, and these parasites also evidenced an attenuated ability to induce contact-dependent cytotoxicity in HeLa cells (
Alderete et al., 1986). Although the significance of viruses in the virulence and pathogenicity of
T. vaginalis remains to be clearly elucidated, it appears that the presence of these viruses or their genomic dsRNA definitely modulated the synthesis and placement of P270 on the surface of the cell. In our pathogenicity test,
T. vaginalis IH-1 and IH-3 evidenced pathogenicity in greater than moderate levels despite the presence of TVV dsRNA. This phenomenon has also been noted in previous reports (
Wang et al., 1987).
T. vaginalis type II isolates which evidenced P270 on the cell surface comprised a heterogeneous population of organisms with and without TVV. When they lost TVV during serial in vitro cultivation, those isolates established a stable phenotype of virulence properties. The same was probably true of the IH-1 and IH-3 isolates. They acquired severe pathogenicity upon long-term cultivation, coupled with a loss of TVV. By way of contrast, the IH-2 isolate evidenced weak pathogenicity, harboring a stable TVV upon prolonged in vitro cultivation (1 year). In the future, a detailed examination of the qualitative and quantitative expression of the surface immunogen, adhesins, and cysteine proteinases will be necessary in order to understand the host-parasite relationship existing between
T. vaginalis and TVV.
The length of the IH-2 cDNA clone (pTVV IH-2) was identical to that of the TVV 1-1 isolate and one base pair short of the TVV 1-5 isolates. Similar conserved motifs at the 5' and 3' termini and an overall sequence of identity of 80% with those of the TVV 1-1 and 1-5 isolates were observed. The same overlapping capsid protein-RdRp gene organization and similar heptamers required for frameshifting were also observed. This evidence showed that the Korean isolate, TVV IH-2, is closely related to TVV 1-1 and 1-5, but is divergent from TVV 2-1 and 3. Although the TVV are currently grouped as tentative members of the
Giardiavirus genus (
ICTVdB Management, 2006), the phylogenetic analysis of RdRp and capsid protein showed that the TVV, and particularly the 1-1, 1-5 and IH-2 isolates, are well separated from GLV, and should be classified as a distinct genera within the
Totiviridae family.
The presence of
T. vaginalis was diagnosed in 22 out of 152 clinical samples (14%), and TVV was detected in 3 of those 22 isolates (14%). The occurrence-ratio of TVV in this study was considerably lower than the previously reported 50% (
Alderete, 1999;
Wendel et al., 2002). There may exist some controversy as to the fact that the samples in this study were collected from patients that had visited hospitals for standard clinical laboratory evaluations, rather than patients who were admitted for testing for sexually transmitted diseases (STD), as was the case in other reports (
Alderete, 1999;
Lawing et al., 2000;
Kaydos et al., 2002;
Wendel et al., 2002;
Alderete et al., 2003). Nonetheless, the presence of TVV or its genomic dsRNA in the Korean IH-2 isolates was significantly associated with the pathogenicity of
T. vaginalis, as has been reported previously.
Notes
-
This work was supported by Inha University Research Grant.
References
Fig. 1The dsRNA profile of TVV IH-2 isolate and its sensitivity to DNase and RNase. Lanes: (M) λ/Hind III DNA marker; (1-3) pBlueScript DNA controls with and without nucleases; (4-7) TVV IH-2 dsRNA with and without nucleases. Arrow on the right indicates the location of dsRNA.

Fig. 2
(A) Genomic organization of TVV IH-2 dsRNA. Two overlapping open reading frames (ORFs) were identified as capsid protein (ORF 1) and dsRNA-dependent RNA polymerase (RdRp, ORF 2), and the overlapping sequence was shown with a potential ribosomal slippage heptamer (bold phase). (B) Sequence context surrounding the putative translation initiation and ribosomal slippage heptamer in the TVV IH-2 isolate. The first Met codon in each of the ORFs was underlined. The overlapping sequence of the 2 ORFs was shown within an open box, and a ribosomal slippage heptamer was indicated in bold. The deduced amino acid sequences of reading frames 1, 2, and 3 in this region were shown in single letter code and the termination codon was designated by an asterisk (*).

Fig. 3The phylogenetic tree determined from the putative RdRp amino acid sequences of the family Totiviridae: Trichomonas vaginalis virus IH-2, 1-1, 1-5, 2-1 and 3 isolates (TVV IH-2, TVV 1-1, TVV 1-5, TVV 2-1 and TVV 3), Saccharomyces cerevisiae virus L-A and L-BC (ScV L-A and ScV L-BC), Giardia lamblia virus (GLV) and Leishmania RNA virus 1-1, 1-4 and 2-1 isolates (LRV 1-1, LRV 1-4 and LRV 2-1). Bootstrap analysis with the unrooted NJ method was conducted using the CLUSTAL W program, and illustrated with the TreeView program. The scaled bar represents distances scaled as substitutions per amino acid residue, and the bootstrap values were indicated at the nodes of each branch.

Table 1.Relationship between pathogenicity of T. vaginalis Korean isolates and the presence of T. vaginalis virus dsRNA
Table 1.
|
Isolate |
No of mice tested |
No of mice with abscess |
Six-day lesions
|
Presence or absence of dsRNA
|
|
Mean volume (mm3) ± SE |
Fresh |
Long term |
|
Control (TYM) |
10 |
0 |
0 |
- |
- |
|
IH-1 |
10 |
3 |
24.91 ± 3.70 |
+ |
- |
|
IH-2 |
10 |
3 |
5.96 ± 2.08 |
+ |
+ |
|
IH-3 |
10 |
8 |
41.48 ± 2.17 |
+ |
- |
|
IH-6 |
10 |
6 |
18.99 ± 3.13 |
- |
- |
|
IH-8 |
10 |
2 |
19.53 ± 1.91 |
- |
- |
|
IH-10 |
10 |
8 |
30.06 ± 4.16 |
- |
- |
|
IH-11 |
10 |
10 |
48.17 ± 8.50 |
- |
- |