Abstract
In a population-based study with 4 years of follow up, we evaluated the prevalence of Coxiella burnetii in cattle on Ulleung Island, Korea. In this study, the rates of C. burnetii infection in cattle on Ulleung Island were determined by PCR and were found to be 0.3–1.0% in the period 2011–2014. All 17 C. burnetii partial 16S rRNA gene sequences from PCR-positive cattle were identical and 2 geographic representatives were included in our analysis. The nucleotide sequences of the 2 samples showed high (98.4–100%) identity with C. burnetii sequences obtained from the GenBank. In this long-term tracking study, the number of cattle positive for C. burnetii on Ulleung Island was low. To prevent the transmission of C. burnetii on Ulleung Island, control strategy should include biosecurity improvement in surveillance, livestock management, administering suitable tests before purchasing animals to detect C. burnetii shedders, and restricting movements between herds.
-
Key words: Coxiella burnetii, cattle, phylogeny, population-based study, 16S rRNA, Ulleung
Q fever is a zoonotic disease with worldwide distribution and is caused by the obligate intracellular bacterium
Coxiella burnetii. Arthropods, mammals, bird, and ticks are reservoirs of infection, and domestic animals, such as cattle, goats, and sheep, are the most significant sources from which the pathogen can be transmitted to humans [
1]. Infection in humans is normally asymptomatic, but can also result in acute or chronic disease [
2]; however, in ruminants, reproductive disorders such as stillbirth, infertility, mastitis, and endometritis are the main clinical symptoms [
3]. The spread of
C. burnetii between ruminant herds can result from transmission between adjacent herds through wind and/or the introduction of infected shedder animals in healthy herds [
4].
In mainland Korea, there have been several serologic and molecular studies on
C. burnetii in cattle [
5–
7], horses [
8], pigs [
9], ticks [
10], dogs [
5], goats [
11], and bulk-tank milk [
6,
12]. Ulleung Island, the second biggest island in Korea, is located 130 km off the east coast of the Korean peninsula. To protect cattle from disease, all cattle on Ulleung Island have been investigated for infectious diseases every year by a government-run local veterinary institute since 2007 [
13]. In our previous enzyme-linked immunosorbent assay (ELISA) study, Ulleung Island was screened for the seroprevalence of
C. burnetii in cattle from 2011 to 2014 [
14]. Serological tests are typically used in epidemiological studies to detect carriers of antibodies against
C. burnetii and show previous exposure to pathogens [
1]. On the contrary, PCR would be more useful to investigate late infection risk of a herd, assuming animal movements happen within the months preceding this changing status [
4]. Therefore, we determined the shedder cattle status for
C. burnetii infection in Ulleung Island with special geographic and epidemiologic situations using PCR.
All cattle from whole farms in Ulleung Island were examined yearly from 2011 to 2014 [
14]. The entire study area was located between the 37°30′0″ north latitude and 131°52′0″ east longitude. Ulleung Island is divided into 3 administrative regions: Ulleung-eup, Seo-myeon, and Buk-myeon. In this study, blood samples were collected from 760 cattle from 54 farms, 597 cattle from 51 farms, 575 cattle from 49 farms, and 625 cattle from 49 farms, each year from 2011 to 2014, respectively. Following the collection of blood, whole blood treated with anticoagulant was used for PCR. Data on the region, age, sex, and breed of cattle sampled were recorded.
Genomic DNA was extracted from blood samples using a commercial DNeasy Blood and Tissue kit (Qiagen, Melbourne, Australia) according to the manufacturer’s instructions and stored at −20°C until use. A commercial AccuPower Hot-Start PCR PreMix kit (Bioneer, Daejeon, Korea) was used for PCR amplification. Multiple primer sets described in previous studies were used to amplify the 16S rRNA gene of the genus
Coxiella, including
C. burnetii and other
Coxiella-like bacteria (CLB) [
7–
10,
12,
14–
16], using nested PCR (nPCR).
Positive PCR products were purified using the QIAquick Gel Extraction kit (Qiagen). Purified products were ligated into a pGEM-T Easy vector (Promega, Madison, Wisconsin, USA), following the manufacturer’s instructions. The ligation product was transformed into Escherichia coli DH5α competent cells and then incubated at 37°C overnight. Plasmid DNA extraction was conducted using a plasmid miniprep kit (Qiagen) following the manufacturer’s instructions.
Recombinant clones in the plasmids were selected and sent to Macrogen (Seoul, Korea) for sequencing. The results were analyzed using the Clustal Omega (ver. 1.2.1) multiple sequence alignment program and the alignment was edited in BioEdit (ver. 7.2.5). The sequence alignment was used to construct a similarity matrix and a phylogenetic analysis was performed using the maximum likelihood method in MEGA (ver. 6.0). The stability of the trees obtained was estimated by a bootstrap analysis with 1,000 replicates.
The chi-square test was used to determine significant differences between multiple groups, with P-values<0.05 regarded as statistically significant. GraphPad Prism ver. 5.04 (GraphPad Software Inc., La Jolla, California, USA) was used for the statistical analyses.
As shown in
Table 1, the rate of
C. burnetii positivity determined by PCR (0.3–1.0%) was low between 2011 and 2014. During the study period,
C. burnetii was found at only 3 farms in the Seo-myeon region, which is what we also observed in our previous seroprevalence study that used the same samples [
14]. Among the 17 PCR-positive cattle of 3 farms from 2011 to 2014, cattle of farms A (1/11, 9.1%), B (1/37, 2.7%), and C (3/56, 5.4%) in 2011, cattle of farms A (1/8, 12.5%) and C (1/57, 1.8%) in 2012, and cattle of farm C in 2013 (6/60, 10%) and 2014 (4/62, 6.5%) showed
C. burnetii infections (data not shown). The Seo-myeon region showed the greatest densities of cattle at both the farm and individual levels compared to those of other regions. Moreover, the 3 farms with
C. burnetii-positive cattle neighbor each other. Although there was no significant trend for
C. burnetii prevalence with respect to both breed and sex, the
C. burnetii prevalence was found to be higher in tiger breed and female cattle than in any other group. Moreover,
C. burnetii positive rates significantly increased with age between 2011 and 2014 (
P <0.05). Upon comparison with the results of our previous ELISA study [
14], 12 out of 17 cattle initially tested positive for
C. burnetii by both PCR and ELISA; however, these 12 cattle eventually became PCR-negative.
In this study, a total of 17 cattle from Ulleung Island were found positive for
C. burnetii infection by PCR between 2011 and 2014. Because all 17 samples contained identical
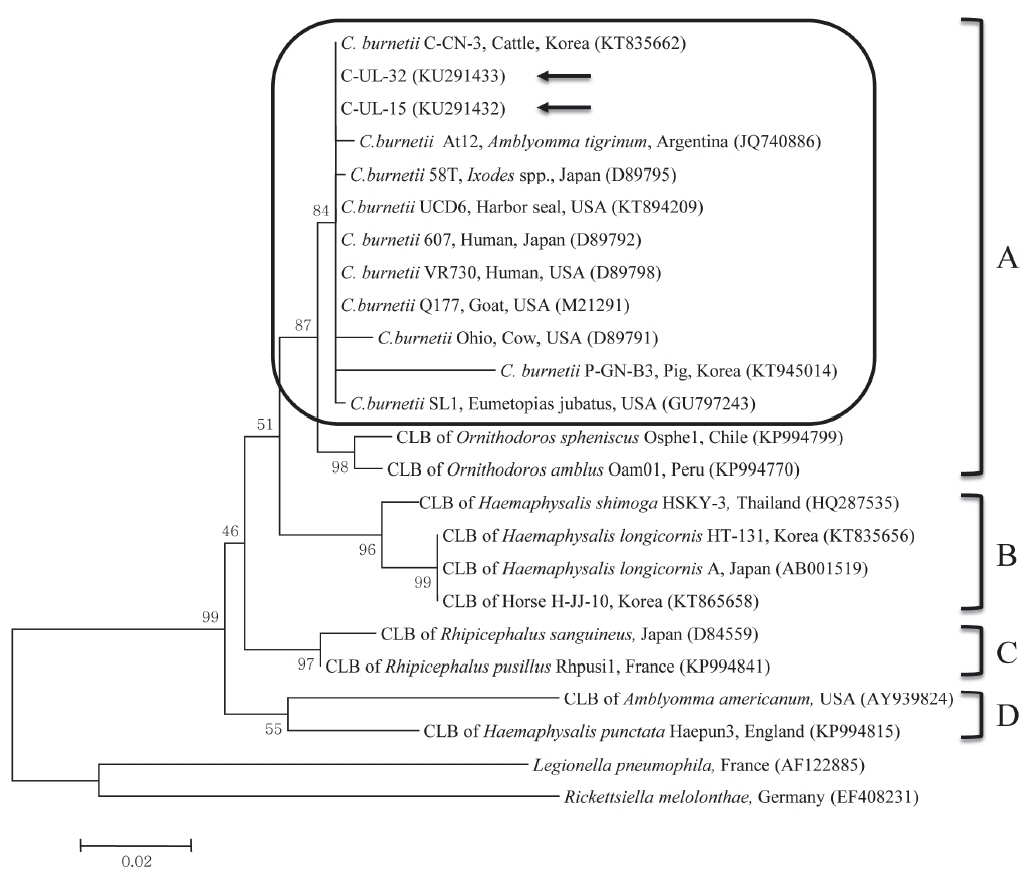
C. burnetii 16S rRNA nucleotide sequences, 2 partial 16S rRNA gene sequences from cattle (C-UL-15 and C-UL-32) in geographically representative areas were deposited in GenBank (accession nos. KU291432 and KU291433, respectively) and used for phylogenetic analysis. Results of the comparative analysis of the 16S rRNA nucleotide sequences from samples C-UL-15 and C-UL-32 and from 19 other bacterial species obtained from GenBank are shown in
Fig. 1. The nucleotide sequences of the 2 samples showed high (98.4–100%) identity with those of other
C. burnetii strains.
We worked on a population-based molecular study with 4 years of follow up on the prevalence of
C. burnetii from cattle in Ulleung Island of Korea. The cattle presented low prevalence of
C. burnetii between 2011 and 2014. Cattle movement from the mainland to Ulleung Island has been limited by the government, resulting in a low risk of
C. burnetii infection for cattle on the island. At the herd level, in small farms like farms A and B with low positive and shedder animals, eradication of
C. burnetii could be reached in a short period of time. The low percentage of shedders observed in these herds suggested a low risk of transmission of
C. burnetii both among herds and from cattle to humans [
17]. However, in larger herds like farm C with active persistent infection and a moderate-to-high percentage of positive animals from 2011 to 2014, more stringent control of
C. burnetii may be needed for a long-term eradication program to protect uninfected animals. We observed the same result in our previous seroprevalence study [
14], such that cattle of farm C showed continuous seroconversions between 2011 and 2014. Because farm C was operated by a government office, farm C had the highest number of breeding cattle on Ulleung Island and frequently had cattle imported from the mainland. Animals in large herds have more chances of transmission between individual animals, increasing the risk of acquiring
C. burnetii infection from imported animals in larger farms [
18]. The previous study also reported that the buying of animals increased the risk of introducing
C. burnetii infection into cattle herds [
19]. Moreover, the detection of multiple animals shedding
C. burnetii DNA in farm C could suggest the progress of the infection, as well as the continuous circulation of
C. burnetii within the herd.
Normally, in isolated islands, there is less movement of cattle between farms. In this regard, in a geographically restricted region like Ulleung Island, restricted movement is important in the prevention and control of diseases. The risk of
C. burnetii infection is influenced both by animal movements and by the proximity of cattle herds. The great effect of neighborhood in high density regions suggests that wind-borne dispersal plays a big role in the increased transmission of
C. burnetii [
4]. In this study, the 3 farms with
C. burnetii-positive cattle were within 1–2 km of each other; thus, these farms may have also been influenced by wind.
C. burnetii positive rates were higher in tiger breed and female cattle, and were found to increase with age in the present study. We observed the same results in our previous ELISA study that used the same samples [
14]. To track the
C. burnetii status in 3 positive farms, additional samples were tested 1 year later (data not shown). Previous positive cattle were already slaughtered, and all farms were negative for
C. burnetii. In this regard, the control strategies to be applied in herds at the regional level may differ according to cattle density and animal movements [
4].
Although the phylogenetic origin of
C. burnetii is unknown, novel CLB that differ from
C. burnetii in their biological characters have been isolated from ticks, suggesting that these CLB may have been symbionts engaged in complex relations with ticks [
16]. Tick-borne
Coxiella strains are known to have widespread genetic variations when compared with the
C. burnetii strains, with
Coxiella being subdivided into 4 divergent clades (A to D) [
16]; although CLB share genetic characteristics with
C. burnetii, their sequences are divergent [
20]. While we assessed the prevalence of
Coxiella, including
C. burnetii and CLB, by targeting the 16S rRNA genes in the present study, we did not amplify CLB genes.
The phylogenetic analysis conducted in this study indicated that the 2 sequences from cattle were closely related to sequences of
C. burnetii strains in clade A. Moreover, the sequences obtained from cattle between 2011 and 2014 were identical, indicating that
C. burnetii strains identified in this study did not change. The 2 sequences analyzed clustered with
C. burnetii isolates from the USA, Japan, and Argentina, which may indicate a close epidemiologic association with these isolates. Although CLB infections were formerly thought to be limited to ticks, a recent study found the first evidence of CLB in horses in Korea, with 0.7% (6/816) positivity based on the sequencing of
Coxiella 16S rRNA gene fragments [
8] using the same primers as those used in this study.
Recently, climate change has affected the distribution of ticks and vector-borne diseases such as Q fever [
21] and, in Korea, the prevalence of ticks and tick-borne diseases has been increasing. For example, ticks on horses have recently been shown to carry CLB (52.4%, 121/213) [
10]; serologic (6.8%, 70/1,030) and PCR (0.3%, 3/1,124) evidences of
C. burnetii infection in pigs have been found [
9]; and dairy cattle bulk-tank milk tested positive for
C. burnetii infection (17.8%, 108/607) by PCR [
12]. Humans exposed to animals and vectors should be evaluated for
C. burnetii infection. Control of coxiellosis in animals is difficult because of the lack of clear clinical signs [
1]. Moreover, farmers often do not recognize the economic significance of Q fever.
Since we could access the whole pattern and features of Q fever distribution in Ulleung Island, we found that the detection of shedders of
C. burnetii was significant and that they are one of the critical points for the control of the spread of the bacteria either among animals or from animals to humans [
2]. Throughout the study period, the infection was shown to be spread and maintained in the animal herd in farm C, while the infection seemed to disappear from farms A and B. Positive farms like farm C should be continuously screened for diseases. Control strategy should include biosecurity improvement in surveillance, livestock management, administering appropriate tests before buying animals to detect assumed
C. burnetii shedders, and restricting movements between herds.
Notes
-
CONFLICT OF INTEREST
We have no conflict of interest related to this work.
Fig. 1Phylogenetic tree of Coxiella partial 16S rRNA gene sequences was constructed using the maximum likelihood method. Coxiella burnetii sequences from this study are marked with arrows. The 4 clades (A to D) of Coxiella are shown and the C. burnetii group in clade A is outlined with a rounded rectangle. The accession numbers of the sequences from GenBank are shown with the sequence names. The numbers on the branches indicate bootstrap support (1,000 replicates). The scale bar represents the evolutionary distance between sequences. CLB, Coxiella-like bacteria.
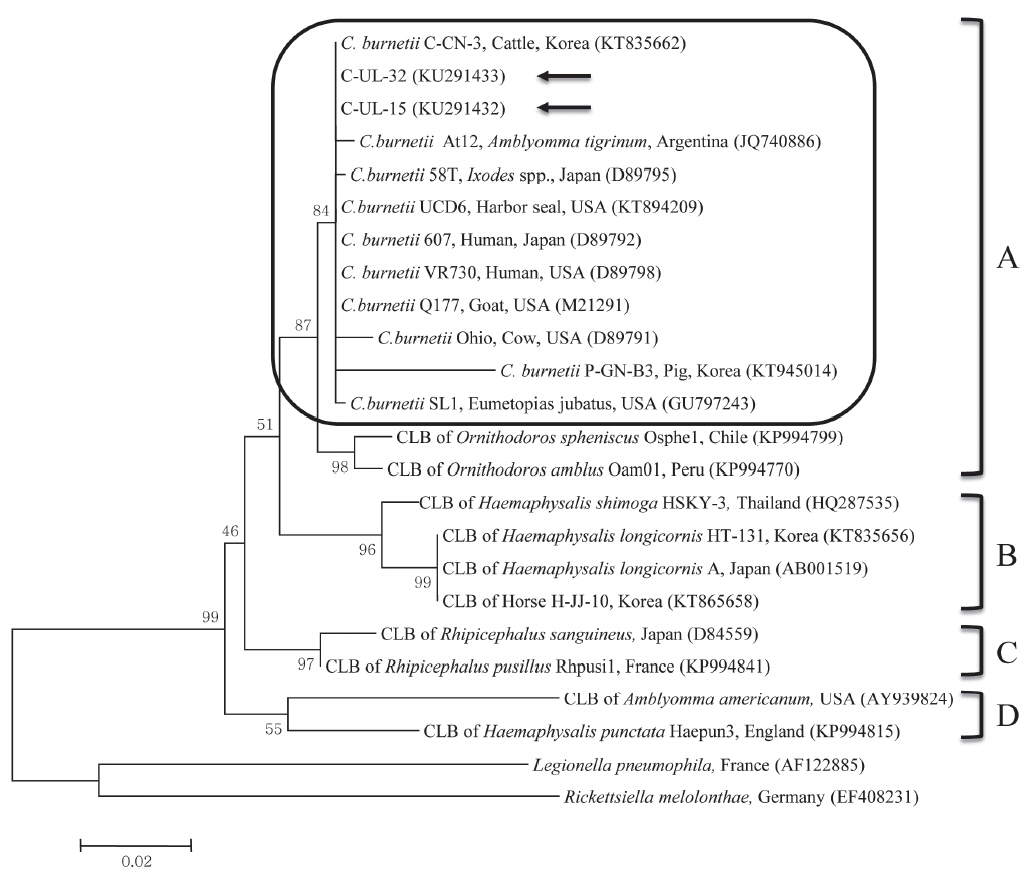
Table 1Prevalence of Coxiella burnetii in cattle reared on Ulleung Island according to breed, sex, and age of cattle, 2011–2014
Table 1
|
Group |
Sub-group |
No. of positive cattle/No. of tested cattle (%) |
|
|
2011 |
2012 |
2013 |
2014 |
|
Breed |
Brown cattle |
1/330 (0.3) |
0/250 |
1/230 (0.4) |
2/323 (0.6) |
|
Tiger cattle |
4/430 (0.9) |
2/347 (0.6) |
5/345 (1.5) |
2/302 (0.7) |
|
P-value |
0.2891 |
0.2292 |
0.2409 |
0.9462 |
|
|
Sex |
Female |
4/431 (0.9) |
2/336 (0.6) |
6/356 (1.7) |
4/370 (1.1) |
|
Male |
1/329 (0.3) |
0/261 |
0/219 |
0/255 |
|
P-value |
0.2916 |
0.2118 |
0.0534 |
0.0958 |
|
|
Age (year) |
<2 |
1/448 (0.2) |
0/305 |
0/237 |
1/336 (0.3) |
|
2–3 |
2/257 (0.8) |
1/256 (0.4) |
0/287 |
0/235 |
|
>3 |
2/55 (3.6) |
1/36 (2.8) |
6/51 (11.8) |
3/54 (5.6) |
|
P-value |
0.0122 |
0.0237 |
<0.0001 |
<0.0001 |
|
|
Total |
|
5/760 (0.7) |
2/597 (0.3) |
6/575 (1.0) |
4/625 (0.6) |
References
- 1. Maurin M, Raoult D. Q fever. Clin Microbiol Rev 1999;12:518-553.
- 2. Arricau-Bouvery N, Rodolakis A. Is Q fever an emerging or reemerging zoonosis? Vet Res 2005;36:327-349.
- 3. Tissot-Dupont H, Raoult D. Q fever. Infect Dis Clin North Am 2008;22:505-514.
- 4. Nusinovici S, Hoch T, Widgren S, Joly A, Lindberg A, Beaudeau F. Relative contributions of neighbourhood and animal movements to Coxiella burnetii infection in dairy cattle herds. Geospat Health 2014;8:471-477.
- 5. Lyoo KS, Kim D, Jang HG, Lee SJ, Park MY, Hahn TW. Prevalence of antibodies against Coxiella burnetii in Korean native cattle, dairy cattle, and dogs in South Korea. Vector Borne Zoonotic Dis 2017;17:213-216.
- 6. Ouh IO, Seo MG, Do JC, Kim IK, Cho MH, Kwak DM. Seroprevalence of Coxiella burnetii in bulk-tank milk and dairy cattle in Gyeongbuk province, Korea. Korean J Vet Serv 2013;36:243-248.
- 7. Seo MG, Ouh IO, Lee SH, Kim JW, Rhee MH, Kwon OD, Kim TH, Kwak D. Prevalence of Coxiella burnetii in cattle at South Korean national breeding stock farms. PLoS One 2017;12:e0177478.
- 8. Seo MG, Lee SH, VanBik D, Ouh IO, Yun SH, Choi E, Park YS, Lee SE, Kim JW, Cho GJ, Kwon OD, Kwak D. Detection and genotyping of Coxiella burnetii and Coxiella-like bacteria in horses in South Korea. PLoS One 2016;11:e0156710.
- 9. Seo MG, Ouh IO, Lee SH, Kwak D. Detection and Genotyping of Coxiella burnetii in Pigs, South Korea, 2014–2015. Emerg Infect Dis 2016;22:2192-2195.
- 10. Seo MG, Lee SH, Ouh IO, Lee GH, Goo YK, Kim S, Kwon OD, Kwak D. Molecular detection and genotyping of Coxiella-like endosymbionts in ticks that infest horses in South Korea. PLoS One 2016;11:e0165784.
- 11. Jung BY, Seo MG, Lee SH, Byun JW, Oem JK, Kwak D. Molecular and serologic detection of Coxiella burnetii in native Korean goats (Capra hircus coreanae). Vet Microbiol 2014;173:152-155.
- 12. Seo MG, Ouh IO, Kwak D. Herd prevalence and genotypes of Coxiella burnetii in dairy cattle bulk tank milk in Gyeongsang provinces of South Korea. Trop Anim Health Prod 2018;50:1399-1404.
- 13. Seo MG, Do JC, Ouh IO, Cho MH, Kim JK, Kim YH, Park NC, Kwak DM. Prevalence of infectious agents in cattle reared in Ulleung island. Korean J Vet Serv 2011;34:303-311.
- 14. Seo MG, Ouh IO, Kim YH, Kim JK, Kwon OD, Kwak D. Serop-revalence of Coxiella burnetii infection in cattle on Ulleung Island, Korea. Korean J Vet Res 2018;58:147-151.
- 15. Duron O, Jourdain E, McCoy KD. Diversity and global distribution of the Coxiella intracellular bacterium in seabird ticks. Ticks Tick Borne Dis 2014;5:557-563.
- 16. Duron O, Noël V, McCoy KD, Bonazzi M, Sidi-Boumedine K, Morel O, Vavre F, Zenner L, Jourdain E, Durand P, Arnathau C, Renaud F, Trape JF, Biguezoton AS, Cremaschi J, Dietrich M, Léger E, Appelgren A, Dupraz M, Gómez-Díaz E, Diatta G, Dayo GK, Adakal H, Zoungrana S, Vial L, Chevillon C. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii
. PLoS Pathog 2015;11:e1004892.
- 17. Pinero A, Barandika JF, Hurtado A, García-Pérez AL. Evaluation of Coxiella burnetii status in dairy cattle herds with bulk-tank milk positive by ELISA and PCR. Transbound Emerg Dis 2014;61:163-168.
- 18. Hogerwerf L, Courcoul A, Klinkenberg D, Beaudeau F, Vergu E, Nielen M. Dairy goat demography and Q fever infection dynamics. Vet Res 2013;44:28.
- 19. J EFSA. European Food Safety Authority, Scientific opinion on Q fever. EFSA Journal 2010;8:1-48.
- 20. Duron O, Sidi-Boumedine K, Rousset E, Moutailler S, Jourdain E. The importance of ticks in Q fever transmission: What has (and has not) been demonstrated? Trends Parasitol 2015;31:536-552.
- 21. Dantas-Torres F. Climate change, biodiversity, ticks and tickborne diseases: The butterfly effect. Int J Parasitol Parasites Wildl 2015;4:452-461.