Abstract
Sinuolinea capsularis Davis, 1917 is myxosporean that infect the urinary system of the host fish. Insufficient morphological and molecular data of S. capsularis exits, and it is therefore difficult to make an accurate identification of the parasite. We tried a series of morphological and molecular analysis to identify an myxosporean isolated from urinary bladder of cultured olive flounder, Paralichthys olivaceus, from Jeju island in the Republic of Korea. Some of them were observed under a light microscope and SEM, and remain samples were used molecular and phylogenetic analysis. Mature spores were subspherical, measuring 13.9±0.6 μm in length and 13.8±0.8 μm in width. Two spherical polar capsules on opposite sides in the middle of the spore had a diameter range of 4.3±0.4 μm. Scanning electron microscopy revealed that spores a severely twisted the suture line. By the morphological comparison and analysis, it was identified as S. capsularis. In addition, we obtained the partial 18S rDNA of S. capsularis and first registered it in NCBI. Phylogenetic analysis showed that S. capsularis clustered with Zschokkella subclade infecting the urinary system of marine fish, and it supported the infection site tropism effect on phylogeny of marine myxosporeans as well as the origin of Sinuolinea is not monophyly.
-
Key words: Sinuolinea capsularis, SEM, phylogeny, olive flounder, PCR
INTRODUCTION
Myxozoans are parasites that infect aquatic organisms and these parasites have a distinct spore stages in vertebrate (mainly fish; myxospore) and invertebrate (mainly annelid; actinospore) [
1]. The parasites have been speciated into 3 main lineages: marine, freshwater, and
Sphaerospora sensu stricto (s.s.) [
2]. Since genus
Sinuolinea was first established by Davis 1917 [
3], approximately 20 species have been reported from marine and freshwater fish [
3–
5]. The parasites are identified by their morphological characteristics, such as spherical or subspherical spores, 2 spherical polar capsules, as well as the sutural line that forms a prominent sinuous ridge around the spore [
3]. Although
S. capsularis was reported in the urinary bladder of
Paralichthys albiguttus, Paralichthys dentatus, and
Spheroides maculates, little information is available for identifying the parasite [
3].
Olive flounder
Paralichthys olivaceus is a prominent fish species cultured in Korea. However, there have been some reports about myxosporean infection in the fish that causing public health and leading to economic loss [
6–
10]. It is important to identify and classify the parasite to control the parasitic disease as well as understand the biodiversity of parasite. Fish Vaccine Research Center has been monitoring the parasitic infections of olive flounder cultured in Jeju Island, and isolated myxosporean spores from the urinary bladder of fish samples. The aim of the present study is to identify the species of myxosporean and reveal phylogenetical characteristics by morphological and molecular analysis. In addition, the dataset obtained from this study will be valuable for future diagnostics and identification of
Sinuolinea spp.
MATERIALS AND METHODS
Parasite samples and partial purification
Olive flounder samples (n=17, 31.4±7.1 cm) were obtained from 4 olive flounder farms situated on Jeju Island. The fish showed extension of urinary bladder with milky urine, and extracts were aseptically obtained using a syringe. The urine suspensions were filtered using a 40 μm cell strainer, followed by centrifugation at 10,000×g (3 min at 20°C). The pellets were resuspended in lysis buffer (RIPA, Merck, Germany) for 5 min, and the suspensions were subsequently centrifuged at 10,000×g (1 min at 20°C). The supernatant was discarded, and the pellet resuspended in phosphate buffer saline (PBS). The sample was collected and preserved at 4°C until further use.
Morphological identification
The urine suspensions and partial purified parasites were wet mounted and observed under a light microscope or a differential interference contrast (DIC) microscope and photographed at 1,000×magnification. The measurements of myxospores were taken from 20 spores each using the ImageJ image processing program (Available from
http://rsb.info.nih.gov/ij/) according to Lom and Arthur’s criteria (1989) [
11].
For scanning electron microscopy, isolated spores were rapidly rinsed twice in PBS and fixed in a solution of 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) at room temperature for 2 hr. The spores were post fixed in 1% osmium tetroxide in the same buffer and dehydrated through ethanol serial solutions of 50, 70, 90, 100, and 100% for 10 min per concentration. Spores were dried in an atmosphere saturated with absolute ethanol, followed by drying with the CO2 critical point method. The samples were sputter coated with platinum, and observed by a JSM-6700F scanning electron microscope at 10 kV, at a working distance 7.8 mm.
Molecular identification and phylogenetic analysis
DNA was extracted from partial purified parasites using AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea) following the manufacturer’s instructions. Portions of 18S rDNA were amplified by PCR using a combination of primers that we (SinuoAKF: 5′-CAWTCYWACTTGGTTRGTTGGTARC-3′, SinuoAKF2: 5′-CYACCTRAGGTTAGCCCATTATWA-3′, SinuoAKR: 5′-ACGGGTTGGTGACCCGTA-3′, and SinuocapR: 5′-CTGCATCCTACTCGGTTCTCA-3′) and other groups (SSU_F04: 5′-GCTTGTCTCAAAGATTAAGCC-3′ and ERIB10 5′-CTTCCGCAGGTTCACCTA-3′) have designed [
12,
13]. PCR was conducted with the following cycling program: initial denaturation of 95°C for 5 min followed by 35 cycles of 95°C for 30 sec, 56°C or 58°C for 30 sec, 72°C for 60 sec or 90 sec, and a final extension at 72°C for 7 min. PCR products were treated with AccuPrep Genomic PCR Purification Kit (Bioneer) to remove excess primers and dNTPs and directly sequenced with BigDyeTM Terminator v3.1 in an ABI 3730xl Sequencer.
Multiple alignments of 18S rDNA sequence were made by Clustal×2.0 [
14] with the homologous sequences of other
Sinuolinea and myxozoans available on the GenBank database. Pairwise sequence distances and similarity of the
Sinuolinea spp. based on 18S rDNA were calculated in MEGA 7.0 [
15] and Clustal Omega [
16]. Bayesian inference (BI) was used to reconstruct the phylogenetic tree from datasets containing 61 sequences of the 18S rDNA from the marine myxosporeans with the malacosporeans, namely
Tetracapsuloides bryosalmonae (KF731712), which was used as an outgroup. For BI analysis, nucleotide substitution models were selected using the Akaike information criterion (AIC), and the Bayesian information criterion (BIC) implemented in jModeltest 2.1.7 [
15,
16] and GTR+I+G and TIM2+I+G were chosen as the best-fit nucleotide substitution models for the 18S rDNA data sets. The metropolis-coupled Markov chain Monte Carlo (MCMC) algorithm implemented in MrBayes 3.2.4 [
19] was run for a sufficient number of generations until the average standard deviation of the split frequencies was<0.05. The sampling frequency was set at every 100 generations for 1,000,000 samples. The first 100,000 samples from each run were discarded as burn-in, and the remaining were analyzed using the “sumt” command in MrBayes. Gaps were treated as missing data. A consensus tree was created using FigTree v1.4.2 (
http://tree.bio.ed.ac.uk/software/figtree/).
RESULTS
We investigated 17 olive flounders and found myxosporean spores in urinary bladder of six fish samples.
Taxonomic summary
Host: Paralichthys olivaceus, Olive flounder (Pleuronectiformes; Paralichthyidae) Locality: Olive flounder culture farm, Jeju Self-Governing Province, Republic of Korea (33°33′N 126°50′E).
Site of infection: Urinary bladder.
Date of sampling: February 2017, January and February 2018.
Type material: Syntype spores had been deposited at the parasitological collection of Fish Vaccine Research Center, Jeju National University Accession Number, PCFVRC20170201A (smear preparation stained with Diff-Quik).
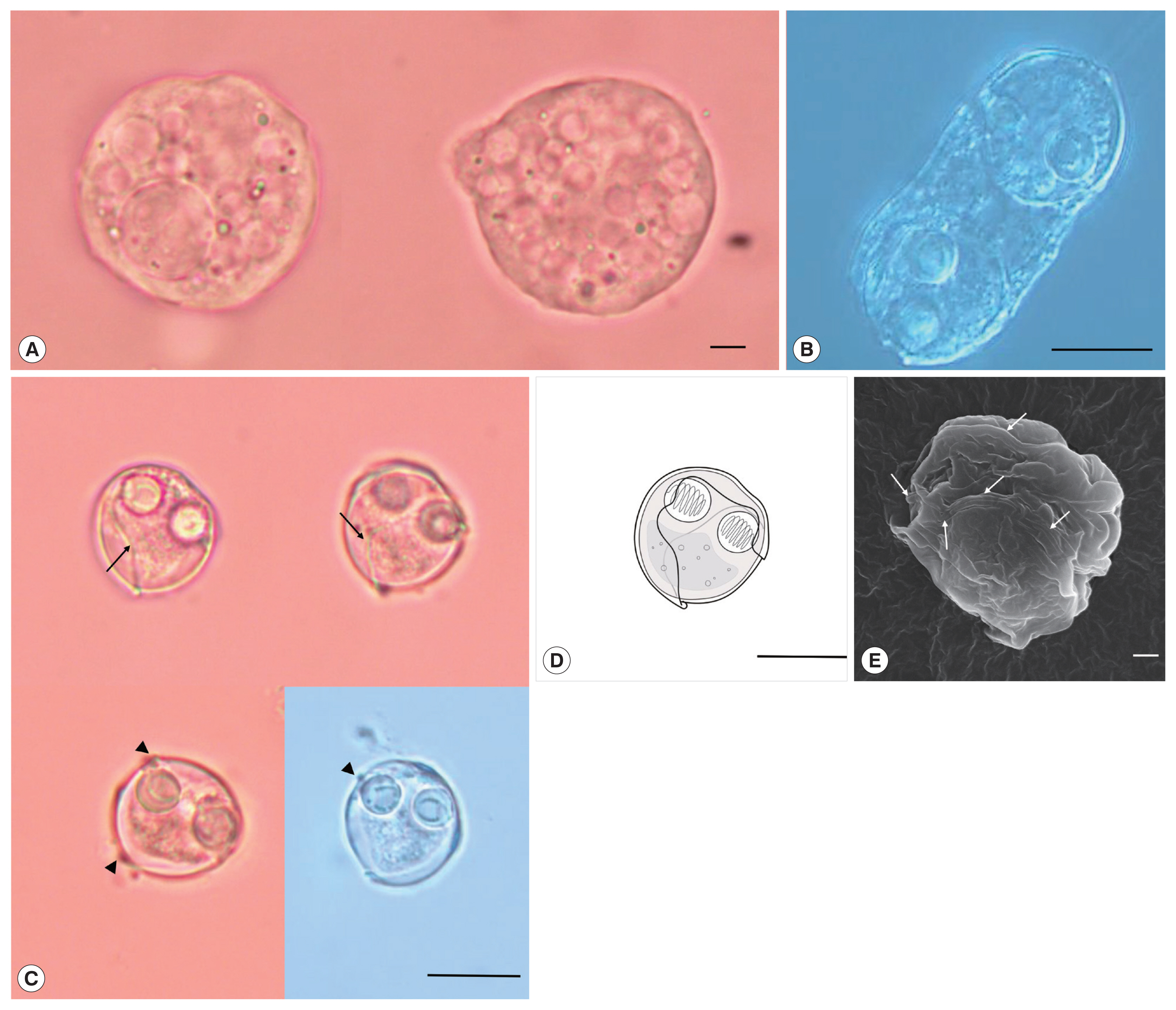
Description: Polysporic plasmodia were found in the urine suspension. They were rough spherical in shape, measuring about 60–70 μm in diameter. The plasmodium had developing spores and 1 large spore such as syncytium, along with a cytoplasmic projection (
Fig. 1A). Disporogonic plasmodia, measuring 25–30 μm in length had 2 spores (
Fig. 1B). The spores were subspherical and measured 13.9±0.6 μm (13.2–15.0 μm) in length and 13.8±0.8 μm (12.7–15.2 μm) in width. Two spherical polar capsules, with a diameter of 4.3±0.4 μm (3.8–5.0 μm), were observed on opposite sides in the middle of the spore. The suture line was twisted and formed a prominent ridge around the spore. The sporoplasm was distinct, with granular globules and polar filament coiled with 5 to 7 turns (
Fig. 1C–E). According to the size of spore and polar capsule, 4 species,
S. capsularis [
3],
S. cyclopterina, [
20,
21],
S. dimorpha [
3], and
S. platycephali [
22] exhibited similarities with the present species.
S. capsularis is most similar with the present species in spore shape and host specificity (Genus
Paralichthys), whereas
S. cyclopterina,
S. dimorpha, and
S. platycephali, have different host specificity (order level) as well as distinct spore shape (
Table 1).
Partial sequences of the 18S rDNA (1,808 bp) was obtained from
S. capsularis and deposited with GenBank (accession number MK072735). A BLAST search indicated that this sequence differed from all available sequences in GenBank as well as the 18S rDNA gene of
Sinuolinea sp. KAB-2001 (AF378346) was the most similar sequences based on the analysis of Max Score. The sequence of the isolate differed from the aligned sequences of all
Sinuolinea species available in Genbank at the 1,554 nucleotide alignments, and had a maximum genetic similarity of 91.5%, being closest to
Sinuolinea sp. KAB-2001 (
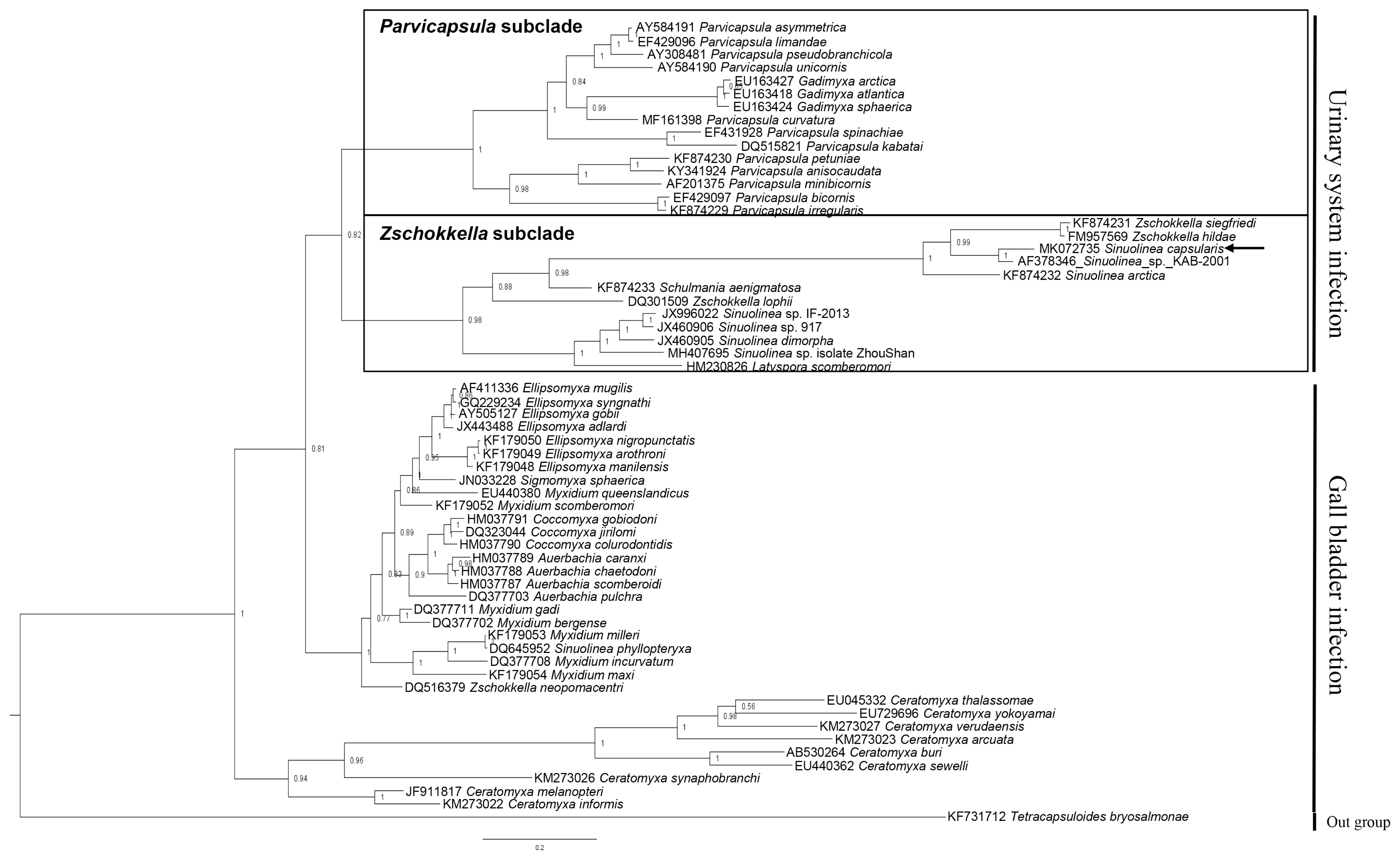
Table 2). The phylogenetic tree constructed by this study divided 2 groups by infection sites (urinary system and gall bladder), and the obtained sequence of
S. capsularis cluster with
Zschokkella subclade in the marine urinary clade (
Fig. 2).
DICUSSION
Davis [
3] explained the shape of
S. capsularis spore to be approximately spherical, sometimes slightly elongated along the longitudinal axis; these features corresponded with our findings. In addition, we observed points with protrusion produced by the sutural line; this is represented in the spore illustration of a previous study [
3]. Sutural line of
S. cyclopterina and
S. dimorpha forming a S-shape; however, other species (including the present species) have a very sinuous and twisted sutural line. The drawing of spore shape and suture line of
S. platycephali reported by Zhao et al. [
22] is similar with
S. capsularis and the present isolate. We could not exclude the possibility that these 3 are the same species (
S. capsularis,
S. platycephali, and present species). However, even if it is proved, they should be identified as
S. capsularis because it is a senior synonym.
Although,
S. capsularis has different hosts (species level) which geographic distribution is limited on the east coast of North America, it is most similar with the parasite isolated from this study based on biological characteristics such as morphology, host (genus level) and infection site. Some of myxosporeans have broader host specificity with infections across families and orders as well as geographical distance [
23–
27].
Kudoa thalassomi has been reported from 18 different fish species representing 6 different fish families and
K. thyrsites has been identified from dozens of marine teleost species in North America, South America, Europe, Africa, Australia and Asia [
23,
25,
26].
Enteromyxum leei also has been found from over 50 fish species in Europe, Africa and Asia including Korea [
9,
24,
27]. In addition, previous study has found
S. capsularis in
Spheroides maculatus (order: Tetraodontiformes) [
3]. Therefore, we identified the present isolate as
S. capsularis, and olive flounder as new host of the parasite.
In our previous study, we revealed the 18S rDNA sequence of
P. curvatura which misidentified as
S. capsularis sequence [
28]. Because the
P. curvatura was not observed in material with light microscopy and we used universal primers (such as 18e, ERIB1 and ERIB10), which have been used to PCR for other myxosporeans [
29,
30], we confused the sequence. Interestingly, Dykova et al. [
31] also reported the similar phenomenon. They obtained the18S rDNA sequences of unidentified Parvicapsulid species that was not observed in light microscopy during investigating the sequence of
Sinuolinea spp. In the present study, we found that the primer 18e and ERIB1 were not suitable to amplify the 18S rDNA sequence in
S. capsularis (data not shown). Thus, we used other universal primer SSU_F04 (used to study on marine metazoan biodiversity and Nematoda) to amplify the 18S rDNA sequence and obtained the sequence of
S. capsularis over 1,800 bp [
13,
32]. We speculate that
S. capsularis has a little different sequence with other myxosporeans in the first forward part of 18S rRNA sequence.
Phylogenetic analysis of myxozoa in previous studies revealed that marine myxosporeans were divided by characteristics of infection site, such as histozoic and coelozoic groups, and the coelozoic group was further divided by infection site as urinary system and gall bladder [
33]. In addition, marine urinary clade consists of
Parvicapsula subclade and
Zschokkella subclade [
34]. Infection site tropism is a strong character effect on the phylogenetic cluster of myxosporeans, and has been reported in previous studies [
35–
38]. In addition, phylogenetic analysis revealed that
Sinuolinea spp. was distributed in
Zschokkella subclade (
S. capsularis,
Sinuolinea sp. KAB-2001,
S. arctica,
Sinuolinea sp. 917,
S. dimorpha, and
Sinuolinea sp. isolate ZhouShan) and in gall bladder infection clade (
S. phyllopteryxa). This finding supports that
Sinuolinea spp. have a para- or polyphyletic character. Previous study also indicated the genus
Sinuolinea did not compose a monophyletic group because
Sinuolinea sequences (JX460905
S. dimorpha, JX460906
Sinuolinea sp. 917) obtained in the previous study had no close relationships with other
Sinuolinea sequences (DQ645952
S. phyllopteryxa, AF378346
Sinuolinea sp.) registered in NCBI [
31].
Other previous studies revealed that phylogeny of myxosporeans have a specific relation with biological traits such as host specificity, infection site tropism, and morphology [
28,
37,
38]. Interestingly, the
Sinuolinea spp. in
Zschokkella subclade were divided into 2 clusters. The
S. capsularis showed the closest genetic similarity with
Sinuolinea sp. KAB-2001 isolated from
Scophthalmus maximus (Pleuronectiformes) in 1 cluster whereas the
Sinuolinea spp. isolated from Perciformes fish were clustered in other part (
Fig. 2;
Table 2). However, it is not enough to claim that there is a specific relation between host specificity and phylogeny of
Sinuolinea spp. because
S. capsularis was isolated from
Sphoeroides maculatus (Tetraodontiformes) as well as the myxosporeans (
S. arctica, Z. siegfriedi, and
Z. hildae) reported from other fish (order level) were also clustered with
S. capsularis. The geographical distance to could be one of the obstacles in deciding the parasite isolated from this study as the same species with
S. capsularis. However, based on the phylogenetic analysis, the geographical distance is not related to genetic similarity in
Sinuolinea spp. The
Sinuolinea sp. KAB-2001 (AF378346) isolated from Spain [
39] was the most genetically close to the present isolate, while
Sinuolinea sp. ZhouShan (MH407695) isolated from China was the most genetically distant to the isolate (
Fig. 2;
Table 2). However, further molecular studies need to clarify the identification of
S. capsularis isolated from the type host and type locality.
Based on the morphological comparisons, the present isolate is identified as S. capsularis. To the best of our knowledge, there is no report about Sinuolinea sp. in olive flounder. Morphological details obtained from the present study suggest the parasite has most similar form with S. capsularis, and the molecular analysis will contribute to reveal its identification, classification and evolution of myxozoan. Unfortunately, there are limited microscopic figure data of Sinuolinea spp. and the sequence of parasites to identify, registered in the NCBI database. Therefore, further morphological and molecular studies need to clarify the identification among Sinuolinea spp.
Notes
-
CONFLICT OF INTEREST
The authors declare no conflict of interest related to this study.
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D1A1B03932598) and is part of the project titled ‘Fish Vaccine Research Center’, funded by the Ministry of Oceans and Fisheries, Korea.
Fig. 1
Sinuolinea capsularis is observed from the urinary bladder of Paralichthys olivaceus. (A) Plasmodium of S. capsularis with developing spores and cytoplasmic projection. Scale bar=10 μm. (B) Disporic plasmodium of S. capsularis. Scale bar=10 μm. (C) Mature spores of S. capsularis with very sinuous sutural line (arrows) and points with protrusion (arrowheads) Scale bar=10 μm. (D) Line drawing of spores of S. capsularis. Scale bar=10 μm. (E) Mature spore of S. capsularis with sutural line (arrows) observed by scanning electron microscope. Scale bar=1 μm.
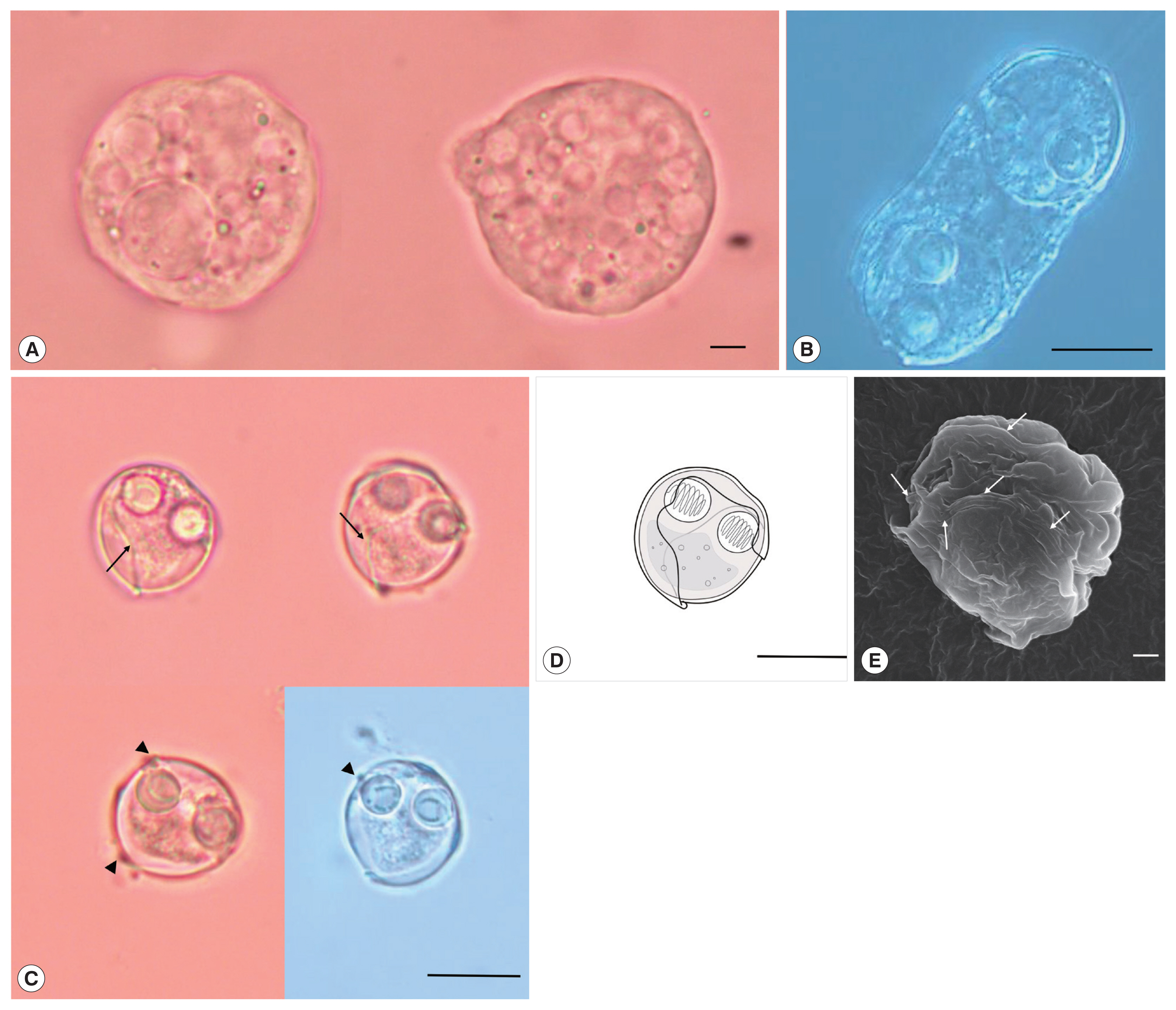
Fig. 2Phylogenetic tree generated by Bayesian analysis (BI) of the aligned partial 18S rDNA sequences of S. capsularis newly obtained from present study and related the marine urinary system infection myxosporeans. The present species were indicated by arrow and clustering with Zschokkella subclade. Tetracapsuloides bryosalmonae was set as out-group and posterior probabilities were listed on the branches.
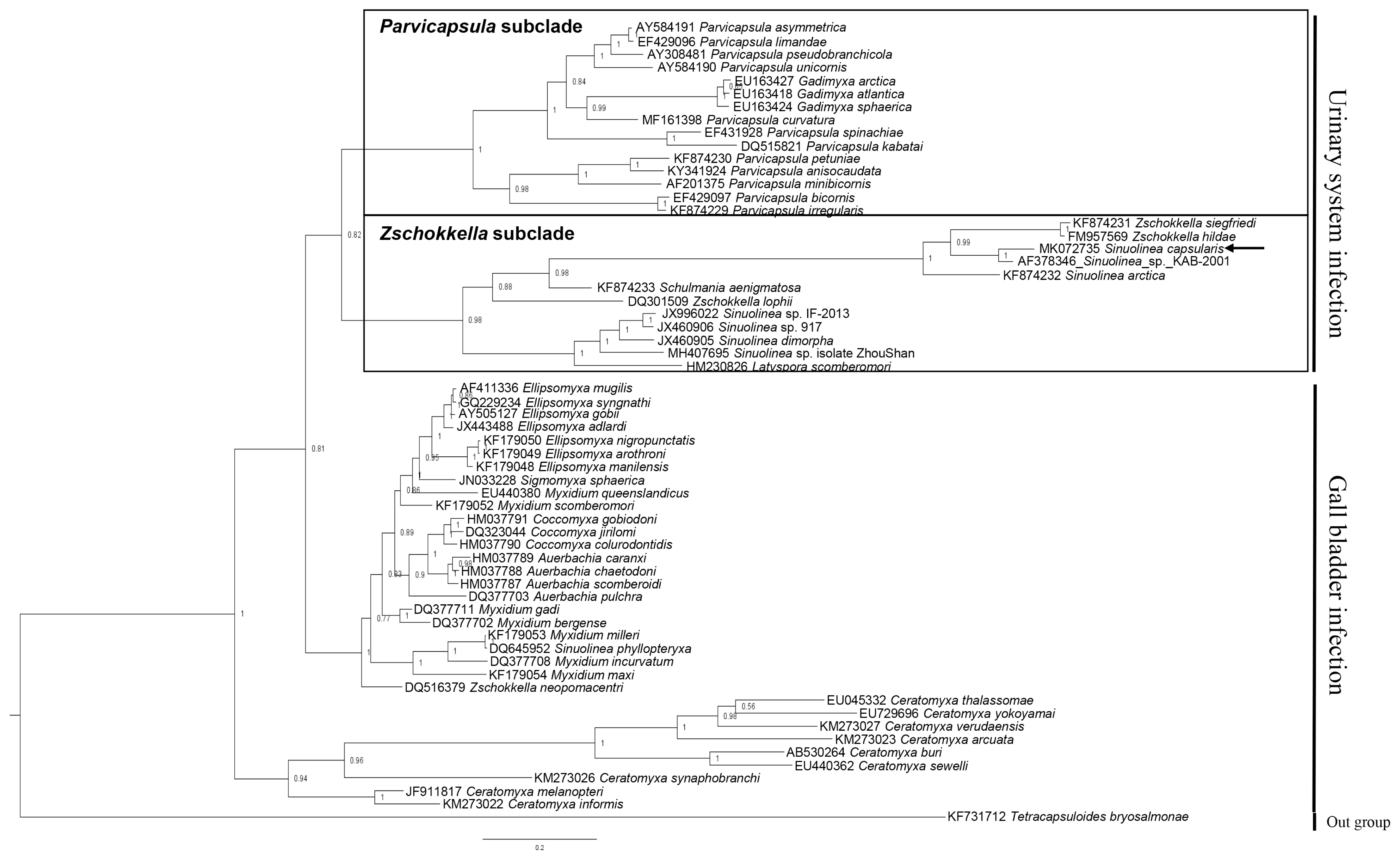
Table 1Comparison on morphology of S. capsularis with other species having similar spore and polar capsule sizes (in μm)
Table 1
|
Species |
S. capsularis
|
S. capsularis
|
S. cyclopterina
|
S. dimorpha
|
S. platycephali
|
|
Host |
Paralichthys olivaceus
|
P. albiguttus, P. dentatus, Spheroides maculates
|
Cyclopterus lumpus
|
Cynoscion regalis
|
Platycephalus indicus
|
|
Spore size |
13.9±0.6 (13.2–15.0) |
12–14 |
14–16 |
15 |
14.5±0.5 |
|
Spore shape |
Subspherical |
Spherical, sometimes slightly elongated |
Spherical |
Spherical |
Spherical |
|
Polar capsule |
4.3±0.4 (3.8–5.0) |
4.5 |
4–5 |
4–5 |
4.3±0.2 |
|
Infection site |
Urinary bladder |
Urinary bladder |
Urinary bladder |
Urinary bladder, ureter |
Urinary bladder |
|
Locality |
Jeju Island, Korea |
Beaufort, USA |
Barents sea, White sea, USSR |
Beaufort, USA |
Yellow sea, China |
|
Reference |
Present study |
[1] |
[18,19] |
[1] |
[20] |
Table 2Genetic distance (P-distance; lower diagonal) and percent sequence similarity (%) (upper diagonal) obtained from the distance matrix based on a 1,554 bp 18S rDNA sequence of Sinuolinea capsularis with all Sinuolinea species available in GenBank
Table 2
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
|
1. MK072735 Present isolate (Sinuolinea capsularis; MK072735) |
|
91.5 |
82.1 |
70.5 |
69.9 |
69.5 |
69.3 |
|
2. Sinuolinea sp. KAB-2001 (AF378346) |
0.05 |
|
80.1 |
70.9 |
68.7 |
68.6 |
68.6 |
|
3. Sinuolinea arctica (KF874232) |
0.16 |
0.16 |
|
69.6 |
67.1 |
68.1 |
67.7 |
|
4. Sinuolinea phyllopteryxa (DQ645952) |
0.32 |
0.33 |
0.34 |
|
80.6 |
80.8 |
80.6 |
|
5. Sinuolinea dimorpha (JX460905) |
0.33 |
0.35 |
0.39 |
0.17 |
|
91.2 |
86.2 |
|
6. Sinuolinea sp. 917 (JX460906) |
0.34 |
0.36 |
0.39 |
0.16 |
0.08 |
|
86.6 |
|
7. Sinuolinea sp. isolate Zhoushan2017 (MH407695) |
0.34 |
0.36 |
0.38 |
0.17 |
0.13 |
0.12 |
|
References
- 1. Lom J, Dyková I. Myxozoan genera: definition and notes on taxonomy, life-cycle terminology and pathogenic species. Folia Parasitol 2006;53:1-36.
- 2. Bartošová P, Fiala I, Jirků M, Cinková M, Caffara M, Fioravanti ML, Atkinson SD, Bartholomew JL, Holzer AS.
Sphaerospora sensu stricto: taxonomy, diversity and evolution of a unique lineage of myxosporeans (Myxozoa). Mol Phylogenet Evol 2013;68:93-105.
- 3. Davis HS. The Myxosporidia of the Beaufort region: a systematic and biologic study. Bull Bur Fish 1917;35:201-243.
- 4. Vaz Rodrigues M, Francisco CJ, Biondi GF, Júnior JP.
Sinuolinea niloticus n. sp., a myxozoan parasite that causes disease in Nile tilapia (Oreochromis niloticus). Parasitol Res 2016;115:4307-4316.
- 5. Garner MM, Atkinson SD, Hallett SL, Bartholomew JL, Nordhausen RW, Reed H, Adams L, Whitaker B. Renal myxozoanosis in weedy sea dragons, Phyllopteryx taeniolatus (Lacepède), caused by Sinuolinea phyllopteryxa n. sp J Fish Dis 2018;31:27-35.
- 6. Kawai T, Sekizuka T, Yahata Y, Kuroda M, Kumeda Y, Iijima Y, Kamata Y, Sugita-Konishi Y, Ohnishi T. Identification of Kudoa septempunctata as the causative agent of novel food poisoning outbreaks in Japan by consumption of Paralichthys olivaceus in raw fish. Clin Infect Dis 2012;54:1046-1052.
- 7. Song JY, Choi JH, Choi HS, Jung SH, Park MA. Monitoring of Kudoa septempunctata in cultured olive flounder and wild fish in Jeju Island during 2012. J Fish Pathol 2013;26:129-137. (in Korean).
- 8. Kim SM, Jun LJ, Park MA, Jeong HD, Jeong JB. Characterization of the myxosporean parasite isolated from emaciated olive flounders Paralichthys olivaceus on Jeju Island. Korean J Fish Aquat Sci 2015;48:337-345. (in Korean).
- 9. Sekiya M, Setsuda A, Sato H, Song K, Han JK, Kim GJ, Yeo IK.
Enteromyxum leei (Myxosporea: Bivalvulida) as the cause of myxosporean emaciation disease of farmed olive flounders (Paralichthys olivaceus) and a turbot (Scophthalmus maximus) on Jeju Island, Korea. Parasitol Res 2016;115:4229-4237.
- 10. Shin SP, Sohn HC, Jin CN, Kang BJ, Lee J. Molecular diagnostics for verifying an etiological agent of emaciation disease in cultured olive flounder Paralichthys olivaceus in Korea. Aquaculture 2018;493:18-25.
- 11. Lom J, Arthur JR. A guideline for the preparation of species descriptions in Myxosporea. J Fish Dis 1989;12:151-156.
- 12. Barta JR, Martin DS, Liberator PA, Dashkevicz M, Anderson JW, Feighner SD, Elbrecht A, Perkins-Barrow A, Jenkins MC, Danforth HD, Ruff MD, Profous-Juchelka H. Phylogenetic relationships among eight Eimeria species infecting domestic fowl inferred using complete small subunit ribosomal DNA sequences. J Parasitol 1997;83:262-271.
- 13. Fonseca VG, Carvalho GR, Sung W, Johnson HF, Power DM, Neill SP, Packer M, Blaxter ML, Lambshead PJ, Thomas WK, Creer S. Second-generation environmental sequencing unmasks marine metazoan biodiversity. Nat Commun 2010;1:98.
- 14. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 2007;23:2947-2948.
- 15. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-1874.
- 16. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011;7:539.
- 17. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003;52:696-704.
- 18. Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 2012;9:772.
- 19. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 2012;61:539-542.
- 20. Basikalowa A. Wissenschaftliches Zentralinstitut für Fischereiwirtschaft. Moscow. 1932, p 136.
- 21. Tripathi YR. Some new Myxosporidia from Plymouth with a proposed new classification of the order. Parasitology 1948;39:110-118.
- 22. Zhao Y, Song W. Studies on the morphology and taxonomy of three new myxosporeans of the genus Sinuolinea Davis, 1917 (Myxosporea: Sinuolineidae) infecting the urinary bladder of some marine fishes from the Shandong coast, China. Syst Parasitol 2003;55:53-59.
- 23. Moran JDW, Whitaker DJ, Kent ML. A review of the myxosporean genus Kudoa Meglitsch, 1947, and its impact on the international aquaculture industry and commercial fisheries. Aquaculture 1999;172:163-196.
- 24. Sitjà-Bobadilla A, Palenzuela O.
Enteromyxum species. In Woo PTK, Buchmann K eds, Fish Parasites: Pathobiology and Protection. Wallingford, UK. CABI; 2012, pp 163-176.
- 25. Whipps CM, Kent ML. Phylogeography of the cosmopolitan marine parasite Kudoa thyrsites (Myxozoa: Myxosporea). J Eukaryot Microbiol 2006;53:364-373.
- 26. Burger MA, Adlard RD. Low host specificity in the Kudoidae (Myxosporea: Multivalvulida) including seventeen new host records for Kudoa thalassomi
. Folia Parasitol 2011;58:1-16.
- 27. Yanagida T. Myxosporean emaciation disease. Fish Pathol 2017;52:63-67.
- 28. Shin SP, Jin CN, Sohn HC, Lee J.
Parvicapsula curvatura n. sp. in cultured olive flounder Paralichthys olivaceus and phylogenetic characteristics of Parvicapsula genus. Dis Aquat Organ 2018;130:199-207.
- 29. Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol 1991;66:411-453.
- 30. Bartosová P, Fiala I, Hypsa V. Concatenated SSU and LSU rDNA data confirm the main evolutionary trends within myxosporeans (Myxozoa: Myxosporea) and provide an effective tool for their molecular phylogenetics. Mol Phylogenet Evol 2009;53:81-93.
- 31. Dyková I, Kodádková A, de Buron I, Fiala I, Roumillat WA.
Sinuolinea infections in the urinary system of Cynoscion species (Sciaenidae) and phylogenetic position of the type species of Sinuolinea Davis, 1917 (Myxozoa: Myxosporea). Int J Parasitol Parasites Wildl 2013;2:10-17.
- 32. Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, Vida JT, Thomas WK. A molecular evolutionary framework for the phylum Nematoda. Nature 1998;392:71-75.
- 33. Fiala I, Hlavničková M, Kodádková A, Freeman MA, Bartošová-Sojková P, Atkinson SD. Evolutionary origin of Ceratonova shasta and phylogeny of the marine myxosporean lineage. Mol Phylogenet Evol 2015;86:75-89.
- 34. Bartošová P, Freeman MA, Yokoyama H, Caffara M, Fiala I. Phylogenetic position of Sphaerospora testicularis and Latyspora scomberomori n. gen. n. sp. (Myxozoa) within the marine urinary clade. Parasitology 2011;138:381-393.
- 35. Holzer AS, Sommerville C, Wootten R. Molecular relationships and phylogeny in a community of myxosporeans and actinosporeans based on their 18S rDNA sequences. Int J Parasitol 2004;34:1099-1111.
- 36. Fiala I. The phylogeny of Myxosporea (Myxozoa) based on small subunit ribosomal RNA gene analysis. Int J Parasitol 2006;36:1521-1534.
- 37. Shin SP, Nguyen VG, Jeong JM, Jun JW, Kim JH, Han JE, Baeck GW, Park SC. The phylogenetic study on Thelohanellus species (Myxosporea) in relation to host specificity and infection site tropism. Mol Phylogenet Evol 2014;72:31-34.
- 38. Shin SP, Shirakashi S, Hamano S, Kato K, Lasso LT, Yokoyama H. Phylogenetic study of the genus Kudoa (Myxozoa: Multivalvulida) with a description of Kudoa rayformis sp. nov. from the trunk muscle of Pacific sierra Scomberomorus sierra
. Mol Phylogenet Evol 2016;98:337-345.
- 39. Kent ML, Andree KB, Bartholomew JL, El-Matbouli M, Desser SS, Devlin RH, Feist SW, Hedrick RP, Hoffmann RW, Khattra J, Hallett SL, Lester RJ, Longshaw M, Palenzeula O, Siddall ME, Xiao C. Recent advances in our knowledge of the Myxozoa. J Eukaryot Microbiol 2001;48:395-413.