Abstract
Rickettsial infections (Rickettsioses) are the causes of acute fever found in Thailand. It is classified as acute febrile illnesses transmitted by bloodsucking arthropod vectors (tick, flea, and chigger). This research investigated pathogens of scrub typhus in vectors from Bangkaew District, Phatthalung Province. A total of 303 pools of vector samples were ticks (Rhipicephalus sanguineus, R. microplus, and Haemaphysalis sp.), fleas (Ctenocephalides felis orientis, C. f. felis, and C. canis), and chiggers (Leptotrombidium deliense, Aschoschoengastia indica, Blankaartia acuscutellaris and Walchia disparunguis pingue) collected from reservoir hosts (dogs and rodents). The 17 and 56 kDa gene of Rickettsia causing scrub typhus were found in 29% of ticks and 98% of flea. DNA sequence analysis reveeled the detected strains were R. asembonensis and Rickettsia sp. cf1 and 5.The chiggers, 1%, were infected with Rickettsia strain TA763, a pathogen of scrub typhus.
-
Key words: Rickettsia, Orientia tsutsugamushi, scrub typhu, bloodsucking arthropod
INTRODUCTION
An infection caused by
Rickettsia (Rickettsioses) is a zoonotic infectious disease caused by bacteria of the family Rickettsiaceae (genus
Rickettsia and genus
Orientia) with blood sucking arthropods (tick, flea and chigger) as vectors and small mammals as reservoir hosts. Humans are also accidental hosts once they are bitten by vector animals that carry the disease. In Thailand, 15–30% acute febrile illnesses (AFI) have been reported to be caused by pathogens of scrub typhus and murine typhus [
1]. Scrub typhus is mostly found in rural areas of Thailand. In 2011, Rodkvamtook et al. reported that 9.8% (17/174) of Thai soldiers in Chonburi province suffered from scrub typhus at the training sites [
2]. According to Rodkvamtook et al. [
3], scrub typhus outbreaks in humans were reported from Mae Rim District, Chiang Mai Province, and the high infection rate was high (86.7%) (26/30). After that, the office of prevention and control 12 (Songkhla) also reported some cases of scrub typhus outbreaks from Bangkaew District, Phatthalung Province. Patient blood extracts were analyzed by Immuno Fluorescent Antibody Test (IFA) to confirm the disease and it was found that 72% (8/11) of the population were infected. This research aimed to isolate the type of blood sucking arthropods and pathogen strains to investigate
Rickettsia and scrub typhus pathogens in arthropod vectors from Bangkaew District, Phatthalung Province.
MATERIALS AND METHODS
Specimen sampling site
During September 2017-January 2018, the specimens of blood sucking arthropods, including ticks, fleas and chiggers, were collected from reservoir hosts (dogs and rodents) from Bangkaew District, Phatthalung Province. Rodents were captured using traps baited with ripe palm and banana. Captured rodents were collected after being euthanized using chloroform. Information of each rodent such as gender, weight, and body length was recorded for species classification. Other specimens (ticks and fleas) were collected from pet dogs. All blood-sucking arthropods were observed and separated to pool of specimens by morphology identification. A pool of specimens included 3 blood-sucking arthropods of same species from same reservoir host. All specimens were preserved in 70% ethanol for preparation of permanent slides and some were stored at −20°C for molecular methods in laboratory.
Genomic DNA extraction and PCR
Genomic DNA of pool of specimens was extracted using GeneJET Viral DNA and RNA Purification Kit (Thermo Fisher Scientific, San Jose, California, USA) according following the manufacturer's protocol and stored at −20°C.
Detection of 17 kDa gene of Rickettsia spp. from tick and flea and 56 kDa gene of O. tsutsugamushi from chigger by Nested PCR
In detection of 17 kDa gene of Rickettsia spp., each PCR reaction mixture was prepared with the following conditions: 4 μl template DNA, 1×PCR buffer (Invitrogen, Carlsbad, California, USA), 2 mM MgCl2, 0.2 mM dNTPs, 1.25 U/μl Taq DNA polymerase and 0.2 μM each forward and reverse primers. The first amplification was carried out using outer primers Rr17.61 (5′-GCTCTTGCAACTTCTATGTT-3′) and Rr17.492 (5′-CATTGTTCGTCAGGTTGGCG-3′). The second amplification was carried out using inner primers F–Jari-17kDa-FDW (5′-CATTACTTGGTTCTCAATTCGGT-3′) and Rr17.492 (5′-CATTGTTCGTCAGGTTGGCG-3′). For detection of 56 kDa gene of O. tsutsugamushi, the first amplification was carried out using outer primers F584 (5′-CAATGTCTGCGTTGTCGTTGC-3′) and RTS9 (5′-ACAGATGCACTATTAGGCAA-3′). The second amplification was carried out using inner primers RTS8 (5′-AGGATTAGAGTGTGGTCCTT-3′) and F (5′-AGCGCTAGGTTTATTAGCAT-3′). The PCR started with an initial denaturation step at 94°C for 3 min, followed by 40 cycles of 94°C for 30 sec, 55°C for 40 sec, and 68°C for 1 min and then a final incubation step of 68°C for 7 min. The amplification product was electrophoresed on 1.5% agarose gels, stained with ethidium bromide and observed under ultraviolet transillumination. The sample was considered to be positive of Rickettsia spp. gene when the 320 bp-specific band and positive of O. tsutsugamushi gene when the 692 bp-specific band was detected.
DNA sequencing and phylogenetic analysis
The PCR products of expected amplicon size were purified using GeneJET Gel Extraction Kit (Thermo Fisher Scientific). The purified products were directly sequenced using dye terminator method (BigDye Terminator sequencing kit, Applied Biosystems, Foster City, California, USA). Consensus sequences were constructed using 3500 Series Data Collection software 3 version 3.1 (Applied Biosystems) followed by removing unexpected gene regions and primer regions. The multi-partial DNA sequences were aligned with reference DNA sequences of genes retrieved from GenBank (
www.ncbi.nlm.nih.gov) using Lasergene DNAstar software (DNAstar Inc., Madison, Wisconsin, USA) using ClustalW algorithm followed by manual modification based on amino acid translation. Nucleotide identity of each DNA sequence was calculated on the basis of pairwise comparison (Lasergene, DNAstar). Neighbor-joining (NJ) were chosen to generate phylogenetic relationships using MEGA 6.0. One thousand bootstrap replicates were applied to all 3 methods.
RESULTS
Rodent and chigger studies
Fifteen rodents were captured from Bangkaew District, Phatthalung Province and could be classified into 5 different species which included 33%
Bandicota indica (5/15), 20%
B. savilei (3/15), 13%
Rattus rattus (2/15), 7%
R. losea (1/15), and 27%
Tupaia glis (4/15) (Armed Forces Research Institute of Medical Science, 2014). The chigger’s morphology was classified according to Nadchaatram and Kethley [
4], resulting in 101 separated specimen pools which included 65%
Leptotrombidium deliense (66/101), 16%
Aschoschoengastia indica (16/101), 18%
Blankaartia acuscutellaris (18/101), and 1%
Walchia disparunguis pingue (1/101). These blood sucking arthropods (101 ticks and 101 fleas) were collected from pet dogs. The morphology of tick, according to Nava et al. [
5], could be classified into 96%
Rhipicephalus sanguineus (97/101), 3%
R. microplus (3/101), and 1%
Haemaphysalis sp. (1/101). The morphology of flea, according to Hiil et al. [
6], could be classified as 97%
Ctenocephalides felis orientis (98/101), 1%
C. f. felis (1/101), and 2%
C. canis (2/101).
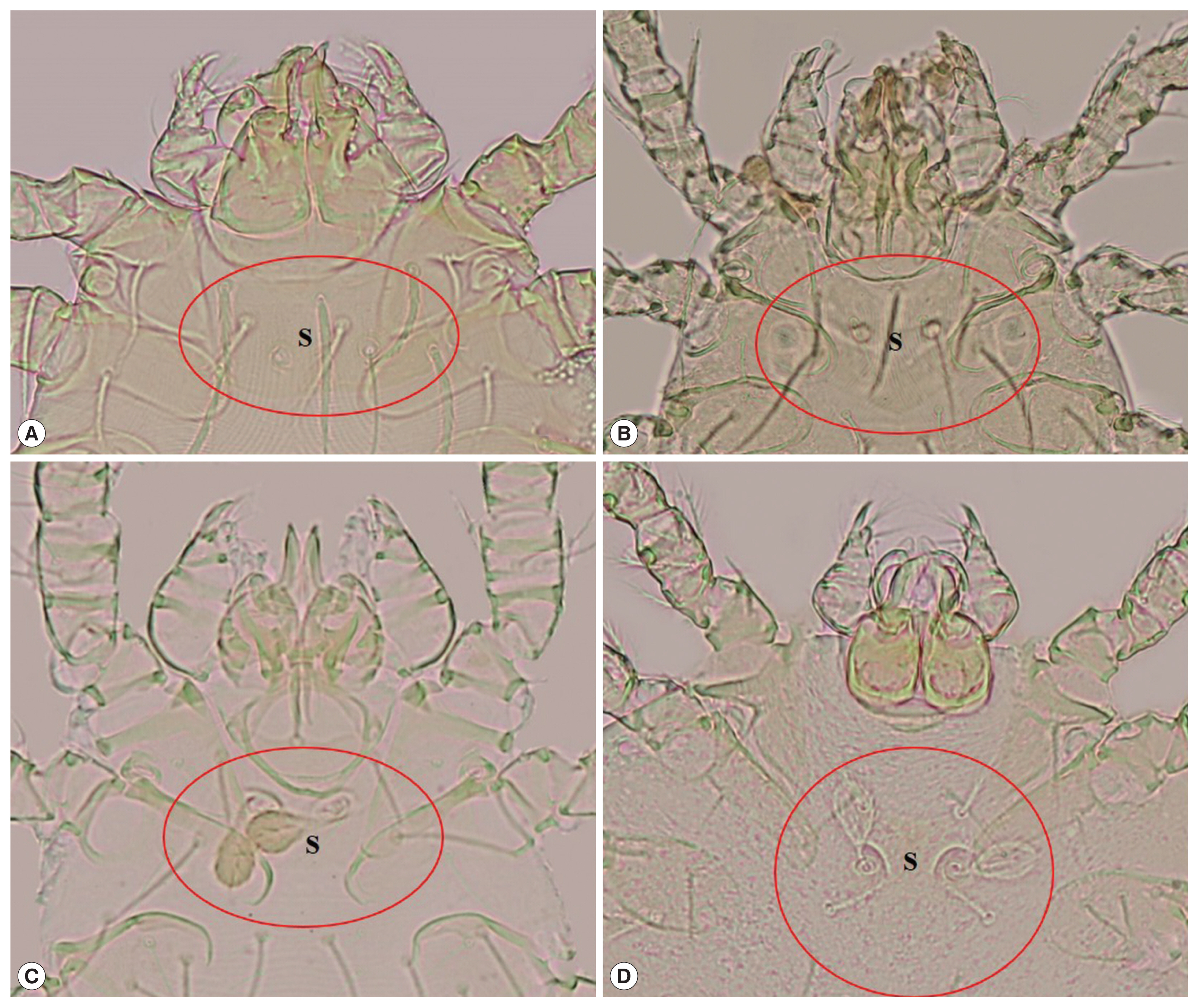
Leptotrombidium delicense: scutum approximately rectangular in shape, with a concave anterior edge, 2 lateral edges and a slightly convex posterior edge, 1 anteromedian setae, 2 pairs of anterolateral setae and posterolateral setae, 2 sensilla on the scutum are filiform with a naked base, formula of dorsal setation=2.8.6.6.4.
Blankaartia acuscutellaris: scutum U or V shape, heavily sclerotized plate, with a concave anterior edge (look like shoulder), all coxae are dash line, arranged in regularity.
Aschoschoengastia indica: scutum lightly punctate, subquadrate with anterolateral shoulders, anterior margin shallowly biconcave, posterior margin convex with median indentation, anteromedian setae base anterior to level of anterolateral seta bases, sensillae setae paddle shape, leg III with nude mastitarsala.
Walchia disparunguis pingue: scutum with 4 scutal setae, antereomedia setae absent, coxa III with 3 setae.
Ticks (Fig. 2)
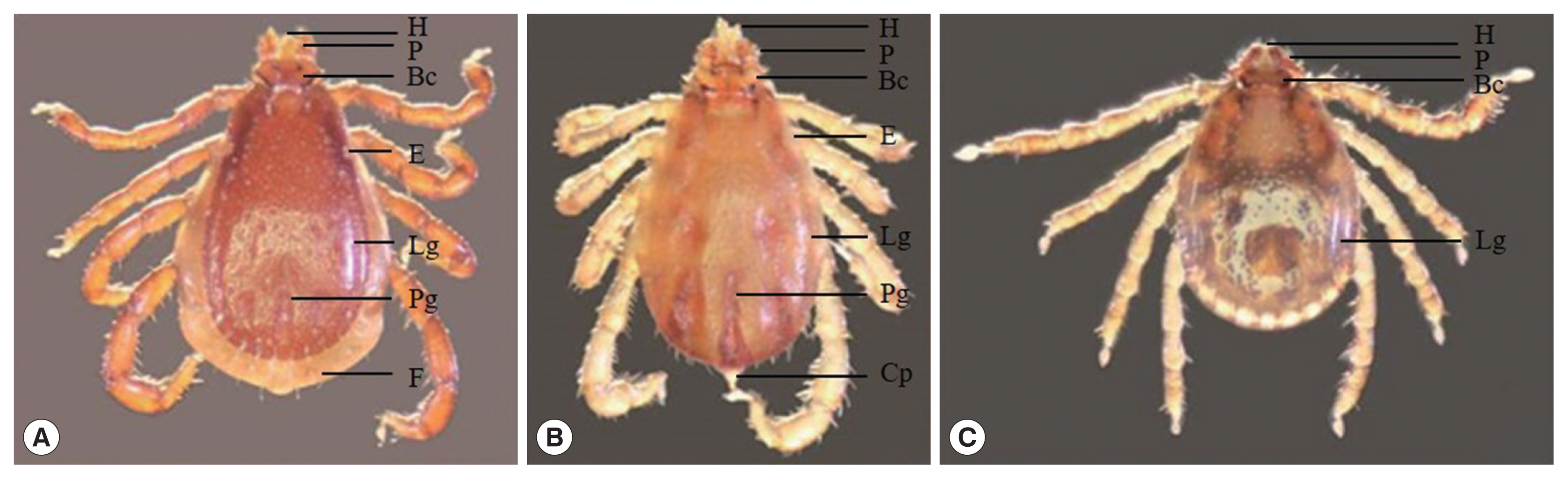
Genus Rhipicephalus: The morphology of genus Rhipicephalus based on the following general characters: eyes present, anal groove posterior to anus, basis capituli hexagonal in shape, palpi short, coxae I deeply cleft, spiracular plates comma-shaped, and male adanal plates and accessory shields present.
Rhipicephalus sanguineus: a pair eye, the palp elongated, larger punctations on the scapular region, basis capituli at about mid-length, posteromedian groove distinctly elongated, lateral grooves circular in shape, festoon present.
R. microplus: a pair eye, the palp and hypostome short and distinctly concave with protuberance absent, alloscutal setae in 2–3 rows, festoon absent, coudal appendage present.
Haemaphysalis sp.: The morphology of genus Haemaphysalis based on the following general characters: the size smaller than Rhipicephalus, eye absent, palp II triangular, distinct lateral grooves and punctations, blade-like dorsal retrograde process on trochanter I, festoon absent.
Fleas (Fig. 3)
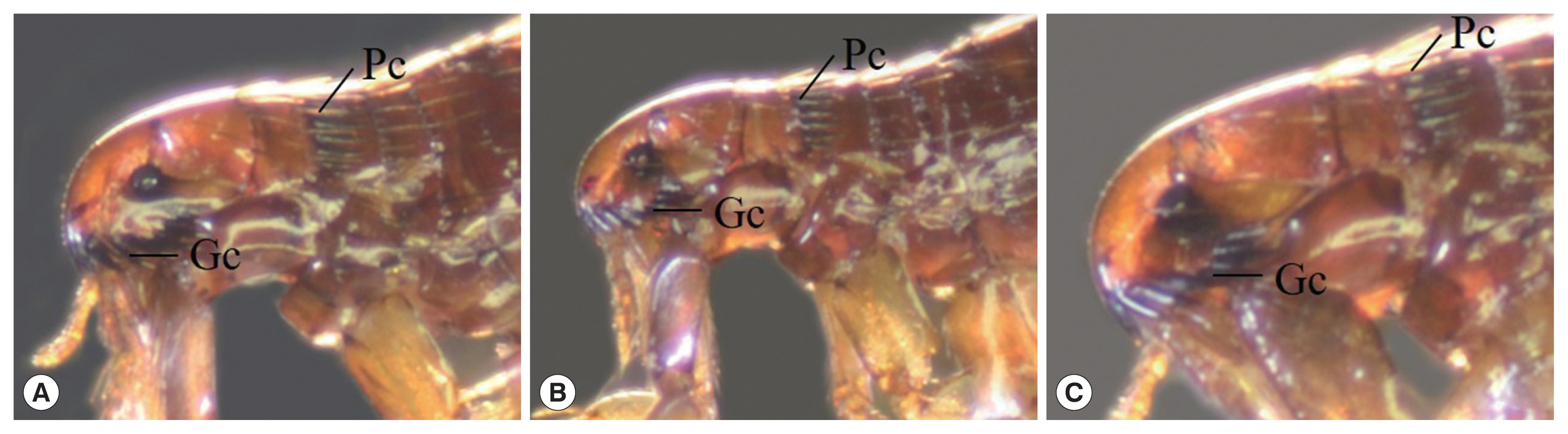
Ctenocephalides felis orientis: anterior genal spine shorter than subsequent spine, the length of head generally greater than twice the height, genal comb I equal in length as next genal comb and consisted of 7–8 pairs of spines, pronotal comb consisted 15–16 spines, tebia with 4–5 pairs of spines.
C. felis felis: the general morphology similar to C. f. orientis but difference on head part of C. f. felis slope down than C. f. orientis.
C. canis: round head shape and the length of head 1.5 times of the height, genal comb I shorter than next genal comb, tebia with 7–8 pairs of spines.
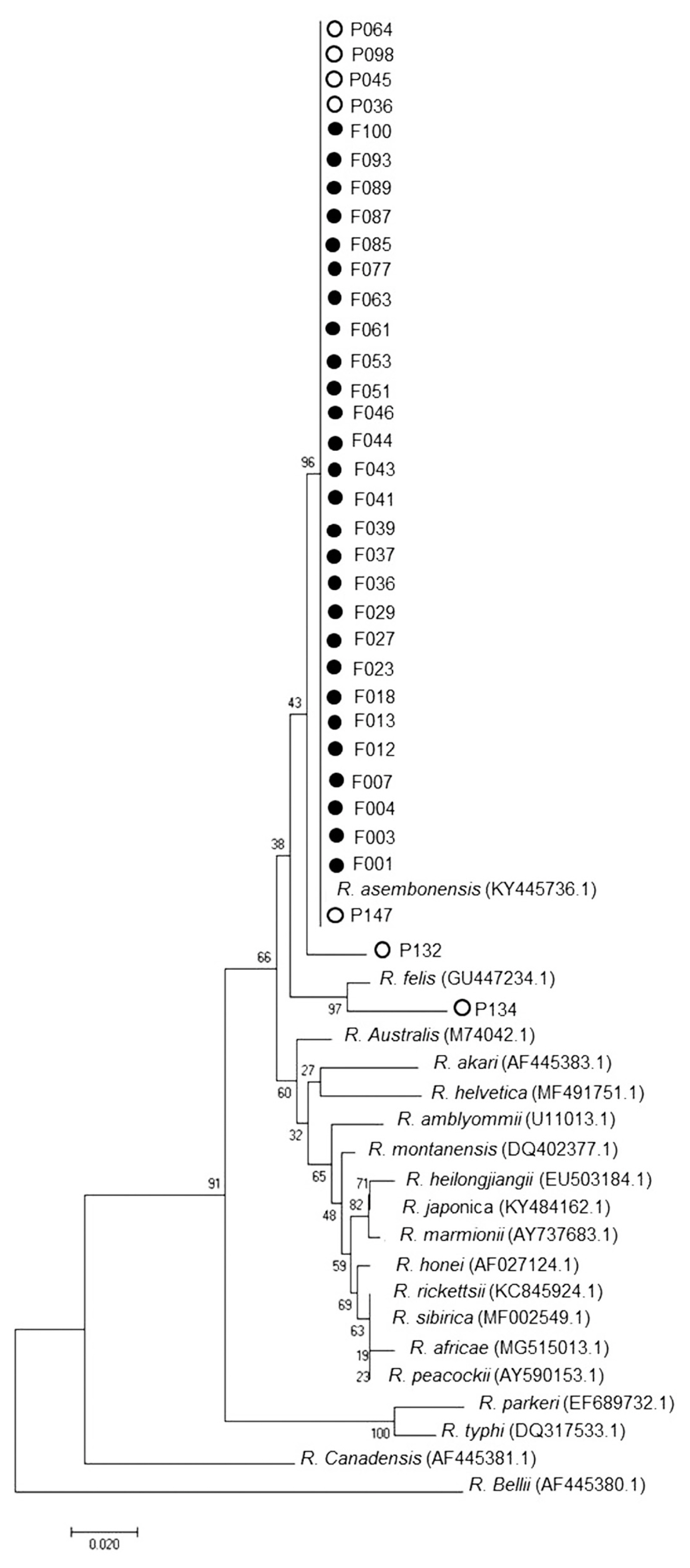
Molecular study
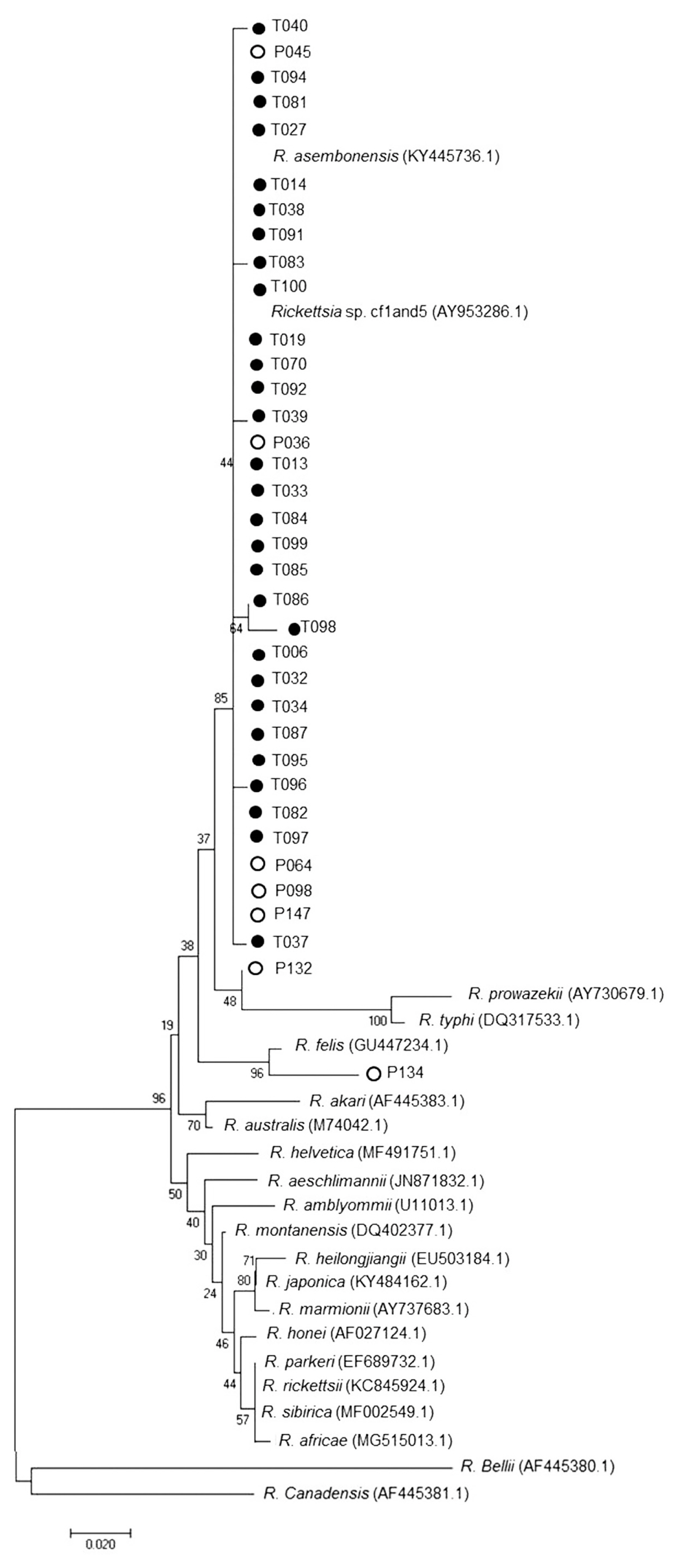
Nested PCR was performed followed by DNA sequence analysis of the 17 kDa gene of
Rickettsia spp. from tick and flea. It was found that 29% ticks (29/101) and 98% fleas (99/101) were positive and could be classified as
Rickettsia spp. which included
R. asembonensis and
Rickettsia sp. cf1 and 5 (using
Rickettsia typhi as positive control and Vero cells as negative control) (
Figs. 4,
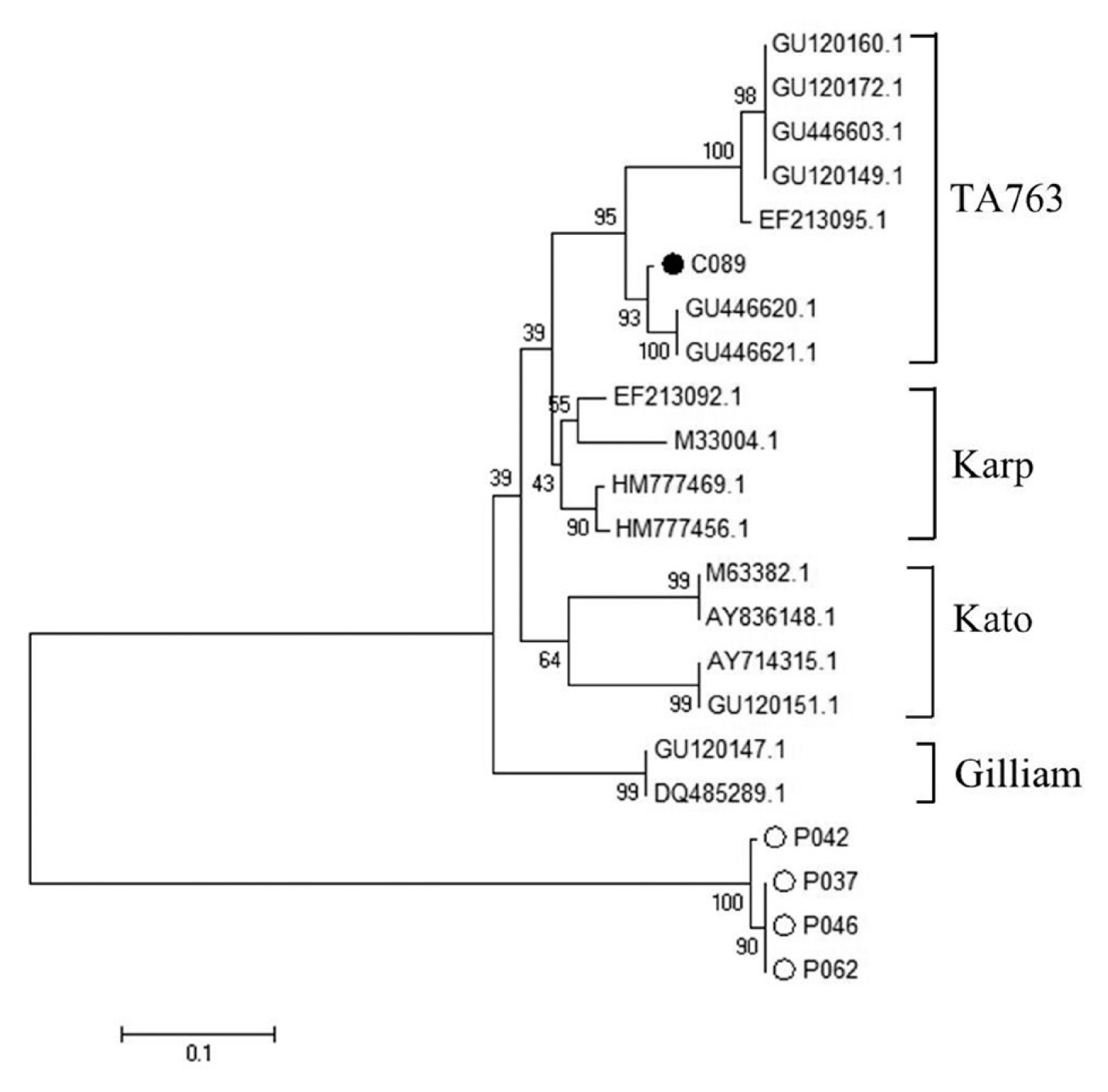
5). The DNA sequence analysis of 56 kDa gene of
O. tsutsugamushi from chiggers found that 1% chiggers (1/101) was positive (using
O. tsutsugamushi strain Karp as positive control and Vero cells as negative control). The phylogenetic analysis was obtained and compared with data from GenBank and it showed that a positive pool was
O. tsutsugamushi strain OI011 or TA763 strain (Thai animal number 763) (
Fig. 6).
DISCUSSION
This study of Rickettsia spp. and O. tsutsugamushi in bloodsucking arthropods (ticks, fleas and chiggers) from Bangkaew District, Phatthalung Province detected 3 species of ticks (Rhipicephalus sanguineus, R. microplus, and Haemaphysalis sp.), 3 species of fleas (Ctenocephalides felis orientis, C. f. felis, and C. canis), and 4 species of chiggers (Leptotrombidium deliense, Aschoschoengastia indica, Blankaartiaacus cutellaris, and Walchia disparunguis pingue). Ticks were infected with R. asembonensis and Rickettsia sp. cf1 and 5. Both stains had not been reported to infect humans while flea could infect but only with R. asembonensis. In chiggers, we found O. tsutsugamushi stain TA763 (Thai animal number 763 or O. tsutsugamushi strain OI011), which had R. rattus as a reservoir host and Leptotrombidium deliense as a vector. However, Thai people live very close to reservoir animals, such as dogs and cats, so they are at increased risk of infection.
The DNA sequence analysis identified 56 pools of tick and flea specimens with positive tests for
Rickettsia spp. Twenty-nine pools of tick specimens were separated to 2 stains which included 68.9%
R. asembonensis and 31.1%
Rickettsia sp. cf1 and 5.
Rhipicephalus sanguineus was the vector of both stains but
R. microplus and
Haemaphysalis sp. were the vector of
R. asembonensis only. The 2
Rickettsia spp. found in this study were not reported to cause diseases in humans, unlike
R. typhi, Murine typhus and
R. felis which were pathogens causing human spotted fever group disease the
Rickettsia sp. that had been diseases reported in humans are [
7]. Kamonwan [
8] reported that
Rickettsia sp. was detected in 7 provinces in Eastern Thailand. They were
Rickettsia sp. Cf1 and 5 that caused infections in
R. sanguineus,
R. asembonensis was not found and
Rickettsia sp. cf1 and 5 had higher infection rates than this study. Moreover, 98% of flea specimens were infected with
R. asembonensis. Phylogenetic relationship analysis with other
Rickettsia found that
R. asembonensis was closely related to
R. felis, as same as the study of Jiang et al. [
9] who reported that, in Kenya,
C. felis and
C. canis were infected with
R. asembonensis and
C. felis was infected with
R. felis. In addition, Maina et al. [
10] had reported that,
R. asembonensis was a new stain and could have worldwide distribution.
Furthermore, DNA sequence analysis of
O. tsutsugamushi from chiggers found that, when compared with GenBank data, one pool of specimen was positive for
O. tsutsugamushi strain OI011 or TA763 (Thai animal number 763, which was similar to the work of Lin et al. [
11] who studied the case in Taiwan but the infection rate (27%, 18/68) was higher than the present study.
Notes
-
CONFLICT OF INTEREST
The authors have no conflicts of interest concerning the work reported in this paper.
ACKNOWLEDGMENTS
Thank you for the research fund from the Graduate school and Biology Program, Faculty of Science, Thaksin University. We would like to specially thank Armed Forces Research Institute of Medical Science (AFRIM) for granting us the use of many of their facilities.
All experimental procedures involving animals were conducted in accordance with the Institute of Animal for Scientific Purposes Development, National Research Council of Thailand (NRCT) (U1-01336-2558).
Fig. 1Chiggers. (A) Leptotrombidium delicense, (B) Blankaartia acuscutellaris, (C) Aschoschoengastia indica, (D) Walchia disparunguis pingue (S: scutum).

Fig. 2Ticks. (A) Rhipicephalus sanguineus, (B) R. microplus, (C) Haemaphysalis sp. (BC: basis capitula, Cp: caudal appendage, E: eye, F: festoon, H: hypostome, Lg: lateral groove, P: palp, Pg: posteromedian groove).

Fig. 3Fleas. (A) Ctenocephalides felis orientis, (B) C. f. felis, (C) C. canis (Gc: genal comb, PC: pronotal comb).

Fig. 4Phylogenetic tree analysis of ticks via Neighbor Joining (NJ) (1,000 bootstrap). Italics words=data from GenBank (NCBI). ●=examples of Rickettsia spp. in this study. ○=Sample of Rickettsia spp. from the blood of patients admitted to Bangkaew Hospital, Bangkaew District, Phatthalung Province (information from Armed Forces Research Institute of Medical Science, Royal Thai Army).

Fig. 5Phylogenetic tree analysis of fleas via Neighbor Joining (NJ) (1,000 bootstrap). Italics words=data from GenBank (NCBI). ●=examples of Rickettsia spp. in this study. ○=Sample of Rickettsia spp. from the blood of patients admitted to Bangkaew Hospital, Bangkaew District, Phatthalung Province (information from Armed Forces Research Institute of Medical Science, Royal Thai Army).

Fig. 6Phylogenetic tree analysis of chiggers via Neighbor Joining (NJ) (1,000 bootstrap). ●=examples of O. tsutsugamushi in this study. ○=Sample of O. tsutsugamushi from the blood of patients admitted to Bangkaew Hospital, Bangkaew District, Phatthalung Province (information from Armed Forces Research Institute of Medical Science, Royal Thai Army).

References
- 1. Thitivichianlert S, Panichkul S, Bodhidatta D, Rodkvamtook W, Sukwit S, Boonmee P, Ketupanya A. Incidence of rickettsial infection in patients with acute fever in provincial Thai army hospitals. J Med Assoc Thai 2009;92( suppl):39-46.
- 2. Rodkvamtook W, Ruang-Areerate T, Gaywee J, Richards AL, Jeamwattanalert P, Bodhidatta D, Sangjun N, Prasartvit A, Jatisatienr A, Jatisatienr C. Isolation and characterization of Orientia tsutsugamushi from rodents captured following a scrub typhus outbreak at a military training base, Bothong district, Chonburi province, central Thailand. Am J Trop Med Hyg 2011;84:599-607.
- 3. Rodkvamtook W, Gaywee J, Kanjanavanit S, Ruangareerate T, Richards AL, Sangjun N, Jeamwattanalert P, Sirisopana N. Scrub typhus outbreak, northern Thailand, 2006–2007. Emerg Infect Dis 2013;19:774-777.
- 4. Nadchaatram M, Kethley J. A collection of reptilian chiggers from Thailand with descritions of three new species (Acari: Prostigmata, Trombiculidae) and preliminary notes on their biologies. J Med Entomol 1974;11:581-587.
- 5. Nava S, Estrada-Peña A, Petney T, Beati L, Labruna MB, Szabó MP, Venzal JM, Mastropaolo M, Mangold AJ, Guglielmone AA. The taxonomic status of Rhipicephalus sanguineus (Latreille, 1806). Vet Parasitol 2015;208:2-8.
- 6. Hiil SF, Lawrence AL, Cuttell L, Tynas R, Rani P, Šlapeta J, Traub RJ. Evidence for a specific host-endosymbiont relationship between ‘Rickettsia sp. genotype RF2125’ and Ctenocephalides felis orientis infesting dogs in India. Parasit Vectors 2015;8:169.
- 7. Parola P.
Rickettsia felis: from a rare disease in the USA to a common cause of fever in sub-Saharan Africa. Clin Microbiol Infection 2011;17:996-1000.
- 8. Kamonwan S. Arthropod vectors for genus Rickettsia and Borrelia in eastern regions of Thailand by Molecular techniques. Master’s thesis. [Bangkok]: Mahidol University; 2015.
- 9. Jiang J, Maina AN, Knobel DL, Cleaveland S, Laudisoit A, Wamburu K, Ogola E, Parola P, Breiman RF, Njenga MK, Richards AL. Molecular detection of Rickettsia felis and Candidatus Rickettsia asemboensis in Fleas from Human Habitats, Asembo, Kenya. Vector Borne Zoonotic Dis 2013;13:550-558.
- 10. Maina AN, Luce-Fedrow A, Omulo S, Hang J, Chan TC, Ade F, Jima DD, Ogola E, Ge H, Breiman RF, Njenga MK, Richards AL. Isolation and characterization of a novel Rickettsia species (Rickettsia asembonensis sp. nov.) obtained from cat fleas (Ctenocephalides felis). Int J Syst Evol Microbiol 2016;66:4512-4517.
- 11. Lin PR, Tsai HP, Tsui PY, Weng MH, Kuo MD, Lin HC, Chen KC, Ji DD, Chu DM, Liu WT. Genetic typing, based on the 56-kilodalton type-specific antigen gene, of Orientia tsutsugamushi strains isolated from chiggers collected from wild-caught rodents in Taiwan. Appl Environ Microbiol 2011;77:3398-3405.