Abstract
Toxoplasma gondii, a ubiquitous, intracellular parasite of the phylum Apicomplexa, infects an estimated one-third of the human population as well as a broad range of warm-blooded animals. We have observed that some tyrosine kinase inhibitors suppressed the growth of T. gondii within host ARPE-10 cells. Among them, afatinib, human epithermal growth factor receptor 2 and 4 (HER2/4) inhibitor, may be used as a therapeutic agent for inhibiting parasite growth with minimal adverse effects on host. In this report, we conducted a proteomic analysis to observe changes in host proteins that were altered via infection with T. gondii and the treatment of HER2/4 inhibitors. Secreting proteins were subjected to a procedure of micor basic reverse phase liquid chromatography, nano-liquid chromatography-mass spectrometry, and ingenuity pathway analysis serially. As a result, the expression level of heterogeneous nuclear ribonucleoprotein K, semaphorin 7A, a GPI membrane anchor, serine/threonine-protein phosphatase 2A, and calpain small subunit 1 proteins were significantly changed, and which were confirmed further by western blot analysis. Changes in various proteins, including these 4 proteins, can be used as a basis for explaining the effects of T. gondii infections and HER2/4 inhibitors.
-
Key words: Toxoplasma gondii, HER2/4 inhibitors, secretome, semaphorin 7A, calpain small subunit 1
INTRODUCTION
Toxoplasma gondii is a member of phylum
Apicomplexa and is the causative agent of toxoplasmosis. It is a widely distributed protozoan parasite that infects various warm-blooded animals, including humans [
1]. It has been reported that 15–85% of various adult human populations show serological evidence of
T. gondii infection, which represents approximately 1/3 of the world population [
2]. Guideline of the Center for Disease Control and Prevention for toxoplasmosis, most healthy people recover from toxoplasmosis without treatment, but at high risk of developing serious diseases if they become immune-compromised or when women get pregnant. Persons who are ill can be treated with a combination of drugs such as pyrimethamine and sulfadiazine with supplement of folinic acid. In case of pregnant women, newborns, and infants can be treated, although the parasite is not eliminated completely. The parasites can remain within tissue cells in a less active phase; their location makes it difficult for the medication to eliminate them. In the case of pyrimethamine and sulfadiazine used for the general treatment of toxoplasmosis, it is difficult to treat toxoplasmosis because it shows side effects called Stevens-Johnson syndrome in some patients [
3].
Decoster et al. [
4] first described several excretory and secretory antigens (ESAs) of
T. gondii in the sera of toxoplasmosis patients. ESA have been studied about cell-mediated immunity [
5–
8], cell biology [
9–
12], and biochemical process [
13,
14]. And Nam et al. [
15] reported about 18 unknown function proteins using proteomic analysis. Nam et al. discovered when
T. gondii infected with host cells, they release various proteins and these large number of secreted proteins have kinase domain. Based on these findings, treatment of kinase inhibitors with
T. gondii infected cells confirmed that it had an anti-toxoplasmosis effect [
16,
17]. These studies suggest the possibility of a vital
T. gondii tyrosine kinase having potential HER2/4 properties, thus anti-HER2/4 tyrosine kinase inhibitors may inhibit intracellular parasite proliferation with minimal adverse effects on host cells.
In this study, we will identify the changes in secreted proteome of host cell through T. gondii (RH) infection and treatment of afatinib. These changes aim to confirm whether kinase inhibitors such as afatinib have potential as therapeutics for toxoplasmosis.
MATERIALS AND METHODS
Ethics statement
All procedures and handling of mice were conducted under an approved protocol by the Institutional Animal Care and Use Committee (IACUC) at the College of Medicine, Catholic University of Korea (CMMC-2015-0042-02, 2015-2017), which adhered to the regulations set under the Korean National Animal Protection Act. The RH strain of T. gondii has been provided by the National Veterinary Research and Quarantine Service.
Cell line and parasite
ARPE-19 cells (ATCC® CRL-2302TM, Manassas, Virginia, USA) were maintained in Dulbecco’s Modified Eagle Medium Nutrient Mixture F-12 (DMEM/F12, Invitrogen, Carlsbad, California, USA) containing 2 μM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml fungizone and 10% fetal bovine serum (FBS, Gibco Life Technologies, Grand Island, New York, USA) (complete DMEM/F12). Tachyzoites of the RH strain of T. gondii were intraperitoneally injected into BALB/c mice, and peritoneal exudates were collected at the 4th day with Dulbecco’s PBS (DPBS).
Drugs and antibodies
Afatinib (BIBW2992) was purchased from Selleck Chemicals (Houston, Texas, USA) as a TKI. DMSO and pyrimethamine were purchased from Sigma Aldrich (St. Louis, Missouri, USA). Afatinib was treated with concentration of 5 mM which obtained over 98% of inhibition of
T. gondii growth within the host cells [
18]. Bovine serum albumin was purchased from Bovogen Biologicals (Melbourne, Australia). FITC-conjugated anti-mouse IgG antibody, TRTIC-conjugated anti-rabbit IgG antibody, and horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibodies were purchased from Sigma Aldrich. Antibody against β-actin was purchased from Cell Signaling Technology (Beverly, Massachusetts, USA). Mouse Tg563 monoclonal antibody was cloned in our laboratory. PDCD4 (D29C6) XP
® anti-rabbit monoclonal antibody was purchased from Cell Signaling (Boston, Massachusetts, USA) Chemicals for the analytical uses as iodoacetamide was purchased from Sigma-Aldrich. Tris-HCl and urea were obtained from Merck (Branchburg, New Jersey, USA). All other chemicals were acquired from standard sources and were of molecular biology grade or higher.
The ARPE-19 cells were cultured in the complete DMEM/F12 (Invitrogen) at 37°C in a humidified 95% air, 5% CO2 incubator. Cells were grown to −70% confluency (−1.6×107 cells) in 150 mm culture dishes (Nunc, Naperville, Illinois, USA). The cell monolayer was rinsed carefully with serum-free medium (SFM) 3 times at RT. Then, the cells were incubated in the SFM at 37°C for 12 hr. After incubation, the SFM from 20 plates was carefully collected with 2 mM PMSF and 1 mM EDTA as protease inhibitors. Floating cells and cellular debris were removed by centrifugation (400 g, 10 min, 4°C), followed by sterile filtration (pore size: 0.22 μm, Millipore, Massachusetts, USA). The conditioned medium was concentrated through ultrafiltration using “Amicon Ultra-15” centrifugal filter devices (Millipore). Secreted proteins were precipitated by acetone at −20°C for 1 hr and then dissolved in buffer consisting of 8 M urea, 75 mM NaCl, 50 mM Tris (pH 8.2). Protein concentration was determined by a standard Bradford protein assay (Bio-Rad, Richmond, California, USA). All protein samples were stored at −80°C until use.
Peptide fractionation by micro bRPLC
The protein sample (100 μg) was reduced with 10 mM DTT (Sigma) at 36°C for 25 min and alkylated with 14 mM iodoacetamide (Sigma) at 25°C for 30 min. The sample was diluted 5 times to decrease urea concentration to less than 1.6 M in solution and added 1 mM CaCl2. The protein mixture was digested by sequencing-grade modified trypsin (Promega, Madison, Wisconsin, USA) at 37°C for 16 hr. The ratio of enzyme to protein was 1:100. Tryptic digests were directly used or further separated based on hydrophobicity by using a Micro bRPLC. Micro bRPLC columns were prepared by adding slurry of 2 mg (10 mg/1 ml acetonitrile) of Jupiter C18 material (5 μm particle diameter, Phenomenex, Torrance, California, USA) to commercially produced microcolumns (C18 Stage Tip™; SP301, ThermoFisher Scientific, West Palm Beach, Florida, USA). All elution steps for column packing, washing, and elution were carried out with benchtop centrifugation (3,000×g for 3 min) unless otherwise stated. Prior to addition of peptide mixtures, the column was washed with 100% acetonitrile (100 μl), then with 100 μl equilibration buffer (100 mM NH4HCO3, pH 8.0). The sample mixture was fractionated with 100 μl portions of 7 different elution buffers (5%, 10%, 15%, 20%, 25%, 30%, and 90% acetonitrile in 100 mM NH4HCO3, pH 8.0).
Protein identification by LC-MS/MS
Nano-high-performance liquid chromatography (nano-HPLC) analyses were performed using an Easy n-LC 1000 system (Thermo Fisher Scientific, San Jose, California, USA). A Q-Exactive mass spectrometer (Thermo Fisher) was used for MS analyses and was operated with Xcalibur (version 2.1) to generate peak lists. The column (30 cm×75 μm) was packed in-house with Jupiter 3 μm, 100 Å pore size C18 beads (Phenomenex). The tryptic peptides were separated using a linear gradient of acetonitrile from 5 to 60% in water in the presence of 0.1% formic acid over a period of 60 min. The mass spectrometer was operated in data-dependent mode with a full scan (m/z 350–2,000) followed by MS/MS for the top 20 precursor ions in each cycle. The acquired MS/MS spectra were subjected to searches against the Uniprot-Human database (June 2018; 73099 sequences) using SEQUEST software in Proteome Discoverer 1.4 (Thermo Fisher Scientific, version 1.4.0.288). Two missed trypsin cleavages were allowed, and the peptide mass tolerances for MS/MS and MS were set to 0.5 Da and 10 ppm, respectively. Other parameters used for the SEQUEST searches included the fixed modification of carbamidomethylation at cysteine (+57.021 Da), the variable modification of oxidation at methionine (+15.995 Da).
Pathway analysis
Proteomic data were uploaded into the Ingenuity Pathways Analysis program (IPA; Ingenuity Systems, Redwood City, California, USA) as a tab-delimited text file of UniGene numbers (transcriptomic data) or IPI accession numbers (proteomic data). Proteins in the proteomic data were mapped to corresponding gene objects in the Ingenuity Pathways Knowledge Base (IPKB). Then, biological networks were generated using the knowledge base for interactions between the uploaded gene list and all other stored gene objects. Functional analysis of the networks was performed to identify the biological functions and/or diseases that were most significant to the genes in the network. For each network, a score was computed according to the fit of the data set of significant genes by Fisher’s exact test. The score was derived from the P-value of the test and indicates the likelihood of the mapped genes in a network being found together due to random chance (score=-log10p).
Western blot
Secretome (10 μg of protein) were size-fractionated by SDS-PAGE and then transferred to a PVDF membrane (Bio-Rad) that was blocked with 5% skim milk in TBS-Tween (25 mM Tris, 190 mM NaCl, 0.05% Tween 20, pH 7.5). The membranes were incubated at 4°C overnight with primary antibodies. Antibodies against RPS12, HNRNPK, PTPRK, SEMA7A, PPP2R2A, RUS1 and CAPNS1 were purchased from Cell signaling. After washing, the membranes were incubated at 25°C for 1 hr with appropriate HRP-conjugated secondary antibodies, washed and then developed with an enhanced chemiluminescence reagent (ECL, GE Healthcare).
RESULTS
Secretome of retinal pigment epithelial cells
To identify new soluble biomarkers that may improve diagnostic tests, we investigated the proteins secreted by
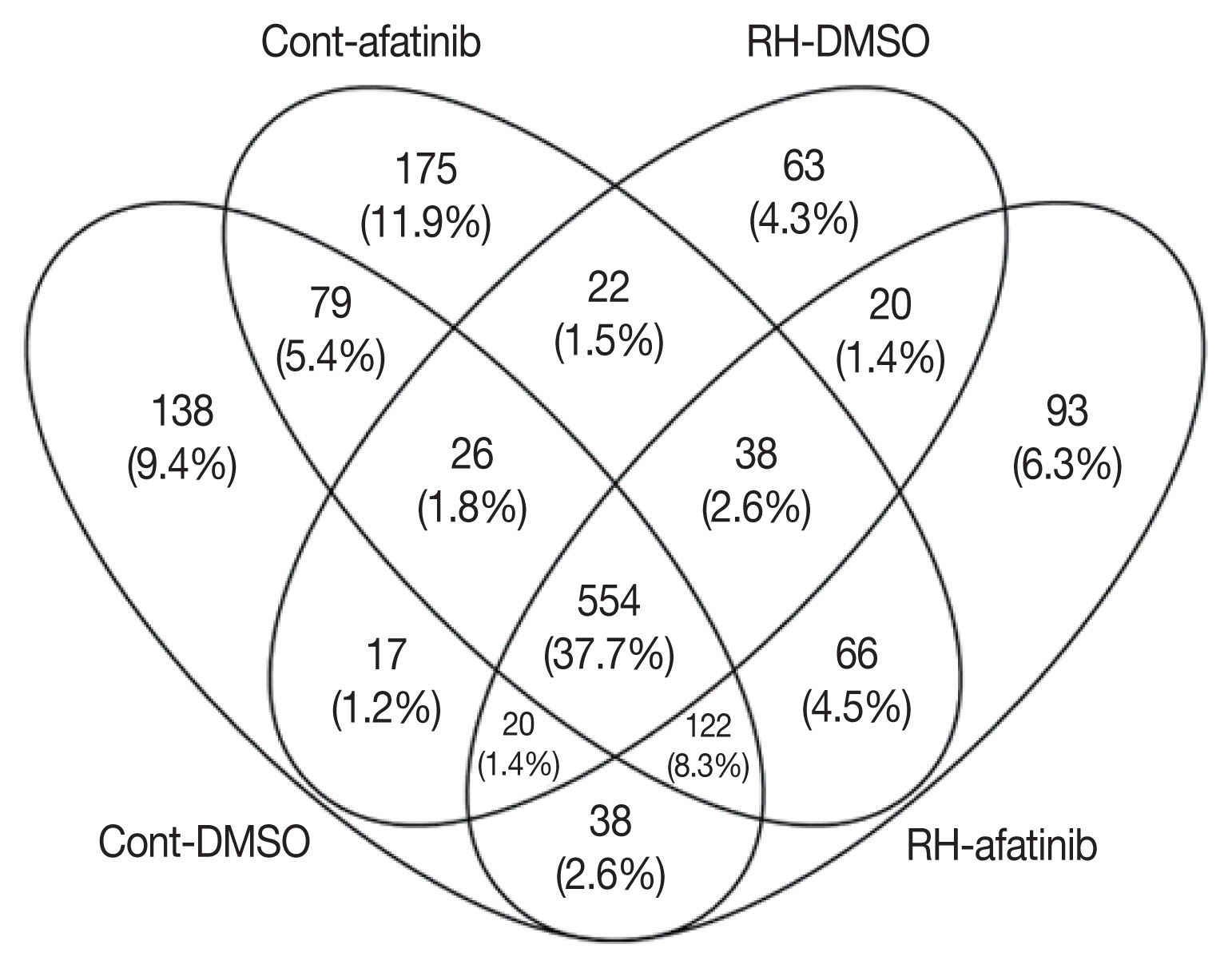
T. gondii using mass spectrometric analyses of conditioned culture media devoid of serum collected from infected ARPE-19 cells. In addition, we performed the secretome analysis of treated-afatinib, which is small molecule of tyrosine kinase inhibitors (TKIs). Overall, 1,867 proteins were identified, and 230 proteins were reported as extracellular space (
Fig. 1), wich showed significant in their fold changes, corresponding p-values, and relevant biological processes.
Ingenuity Pathways Analysis programs were used to predict the subcellular localization of the identified proteins and their biological functions. The searched proteins were linked to at least one annotation terms. The distribution of subcellular localizations was cytoplasm (46%), Cytosol (11%), extracellular space (10%), unknown (8%), cell surface (7%) and membrane (5%). Gene Ontology analysis showed that these proteins were mainly involved in protein synthesis and cell death and survival.
To put our proteomics data in a biological context, the expression data were analyzed using IPA software. All proteins were mapped to gene objects in the Ingenuity Pathways Knowledge Base. To further understand the changes observed in the secretome upon
T. gondii infection, 269 DEPs were analyzed using IPA software. Eighteen networks were generated from the 269 DEPs. The 3 networks were merged to obtain a global view of the proteins that were differentially regulated between the T. gondii-infected and–uninfected secretome. To further understand the changes observed in the secretome upon treated-Afatinib, 168 DEPs were analylyzed using IPA software. Eleven networks were generated from the 168 DEPs. The 3 networks were merged to obtain a global view of the proteins that were differentially regulated between the treated-Afatinib and–untreated secretome. Interestingly, there were some major hubs in the network. AKT, which has been shown to play key roles at tumor progression and apoptosis, is connected to several DEPs along with the ERK/MAP kinase protein family. Further, there are 15 known drug targets in cancer ERK, p38 mitogen-activated protein kinase (MAPK), Cytokeratin, keratin, HDL-cholesterol, SAA, DNA-pK, MTORCs, CDK4/6, AKT, MTR, Cadherin, 60S ribosomal subunit, PRKAA, Cyclin D) that are a part of the network. The list of common protein in global network is shown in
Table 1. In the next step, the 230 extracellular proteins were mapped onto known canonical signaling pathways in order to obtain useful information on molecular interaction networks. The top canonical signaling pathway was acute phase response signaling, which was populated with 18 proteins from the network. The list of acute phase response signaling for extracellular protein is shown in
Table 2.
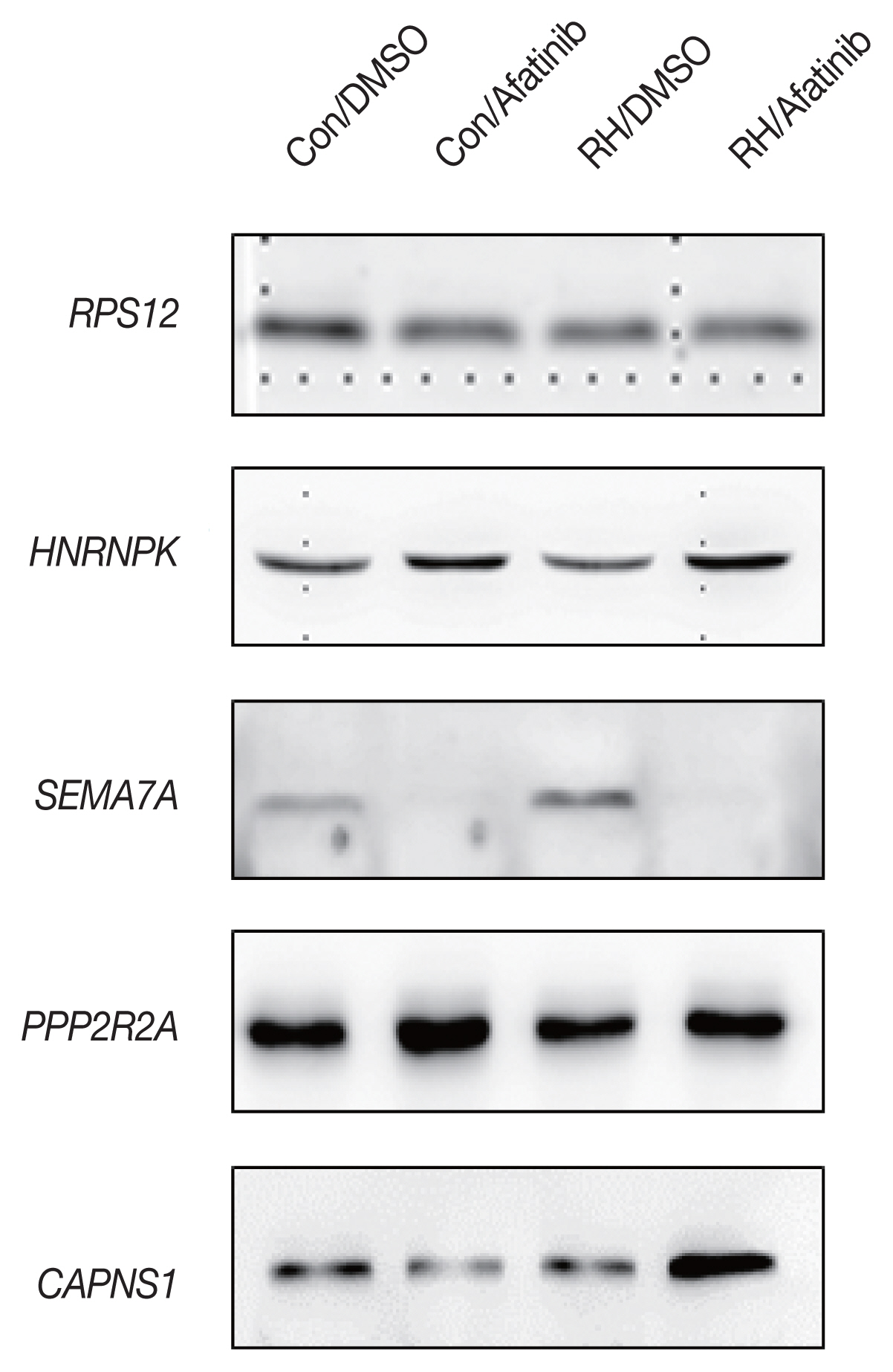
The initial 7 candidate proteins were narrowed down with regard to their biological functions drawn from IPA and the availability of commercial antibodies. Out of 7 proteins, only 4 showed significantly different concentrations between the groups: HNRNPK (Heterogeneous nuclear ribonucleoprotein K, hnRNP K), SEMA7A (Semaphorin-7A), PPP2R2A (Serine/threonine-protein phosphatase 2A) and CAPNS1 (Calpain small subunit 1) (
Fig. 2).
DISCUSSION
Despite great effort and much progress, toxoplasmosis remains a major threat to global health. To identify new soluble biomarkers that may improve diagnostic tests, we investigated the proteins secreted by T. gondii using mass spectrometric analyses of conditioned culture media devoid of serum collected from infected ARPE-19 cells. In addition, we performed the secretome analysis of treated-Afatinib, which is small molecule of HER2/4 inhibitor using anti-cancer drug.
Overall, 1,867 proteins were identified, and 230 proteins were reported as extracellular space. Gene Ontology (GO) analysis showed that these proteins were mainly involved in immune response signaling and cell death and survival. These results provide new insight into the essential regulators of host-pathogen interactions. Looking at the results of these complex protein network analyzes, we found quite interesting results (
Table 1,
2). In the acute phase response signaling, the expression pattern of protein changed through
T. gondii infection appears to be the opposite of the expression pattern of protein changed by afatinib treatment. TTR, SERPING1, SERPINF1, SERPINF2, SERPIND1, SERPINA3, RBP4, ITIH2, ITIH3, IL6, IL18, HPX, C4A/C4B, C1S, C1R, APOH, ALB, and A2M are examples of proteins with decreasing or increasing plasma concentration in acute phase response signaling. One of the most prominent of these proteins is SERPIN, which controls an array of biological processes, including coagulation and inflammation, and consequently these proteins are the target of medical research [
19,
20]. It is difficult to deduce whether the increase or decrease in plasma of these proteins is simply the result of host cell reaction by afatinib or how afatinib affects RH and is the result. However, these changes may be an important first step in explaining the treatment of
T. gondii caused by HER2/4 inhibitors such as afatinib.
Several proteins capable of securing antibodies were selected, and 4 proteins showed significant changes through infection with
T. gondii and treatment with afatinib through Western blot analysis. In the case of HNRNPK, a gene that is directly involved in the transcription of a host cell, particularly hnRNPK, which is important for inducing apoptosis [
21,
22]. Western blot results show that RH infection lowers HNRNPK expression level and afatibib treatment increases HNRNPK expression level. Inferred from reports that low levels of HNRNPK protein affect apoptosis, RH infection interferes with apoptosis and afatinib increases HNRNPK expression to induce apoptosis to eliminate RH-infected cells.
SEMA7A have an important role in integrin-mediated signaling and functions both in regulating cell migration and immune responses [
23]. And it Plays an important role in modulating inflammation and T-cell-mediated immune responses [
24,
25]. The results suggest that afatinib significantly reduces the expression of SEMA7A. If these results indicate that the therapeutic effect of
T. gondii worms is significant, then a closer look at the pathway through the protein is required.
PPP2RA is a protein that regulates the function of PPIA (Peptidyl-prolyl cis-trans isomerase A) [
26]. It is a protein that regulates conformation of proteins or RNA binding, which does not seem to be directly related to
T. gondii. However, the Biological process of GO database related to PPP2RA is related to the invasion of symbiotic organisms targeting host cells. In other words, PPP2RA is estimated to be important for RH infection of host cells and life cycle. In Western blot analysis results, afatinib increases the expression level of intracellular PPP2RA. This change in expression seems to affect the survival of RH.
CAPNS1 is regulatory subunit of the calcium-regulated non-lysosomal thiol-protease which catalyzes limited proteolysis of substrates involved in cytoskeletal remodeling and signal transduction [
27,
28]. Interestingly, the levels of CAPNS1 are significantly higher when afatinib is treated in toxic RH-infected cells.
Although these changes do not directly explain the effects of afatinib on RH-infected hosts, it is important to note the possibility that they may be one of the key molecules in the treatment of T. gondii. Functional characterization of the altered proteins may enhance understanding of the host responses to T. gondii infection and lead to the identification of new therapeutic targets.
Notes
-
CONFLICT OF INTEREST
There are no conflicts of interest to declare.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2017RIA2B4004594).
Fig. 1Proteomic feature of the ARP-19 cells infected with RH strain, and treated with afatinib.

Fig. 2Western blot of the selected proteins from each subgroup by MS analysis. All expression levels were normalized to RPS12 (40S risosomal protein S12). HNRNPK: Heterogeneous nuclear ribonucleoprotein K; SEMA7A: Semaphorin 7A; PPP2R2A: PP2A subunit B isoform B-55-alpha; CAPNS1: Calpain small subunit 1.
Table 1Protein network analysed using Ingenuity Pathway Analysis
Table 1
|
Entrez gene name |
Fold changea
|
|
Bone morphogenetic protein 1 |
0.5 |
|
c-type lectin domain family 3 member B |
−0.3 |
|
collagen type XVIII alpha 1 chain |
2.9 |
|
Collagen type V alpha 1 chain |
2.9 |
|
Cellular repressor of E1A stimulated genes 1 |
3.1 |
|
cystatin B |
−0.9 |
|
cathepsin B |
2.5 |
|
cathepsin L |
2.7 |
|
EGF containing fibulin extracellular matrix protein 2 |
2.2 |
|
Fibulin 1 |
2.4 |
|
fibromodulin |
3.1 |
|
Galactosidase beta 1 |
−2.3 |
|
Hornerin |
−1.5 |
|
Inter-alpha-trypsin inhibitor heavy chain 2 |
3.2 |
|
Inter-alpha-trypsin inhibitor heavy chain 3 |
3.6 |
|
Laminin subunit gamma 1 |
3.5 |
|
Lysyl oxidase |
2.6 |
|
Pentraxin 3 |
3.6 |
|
S100 calcium binding protein A11 |
−2.3 |
|
Secretogranin II |
4.2 |
|
Serpin family G member 1 |
4.9 |
|
Thrombospondin 4 |
−2.2 |
|
Tolloid like 1 |
2.4 |
|
TNF alpha induced protein 6 |
2.4 |
Table 2Acute phase response signaling for extracellular proteins
Table 2
|
Entrez gene name |
Fold changea
|
|
Transthyretin |
2.4 |
|
Serpin family G member 1 |
4.9 |
|
Serpin family F member 2 |
10.7 |
|
Serpin family F member 1 |
2.3 |
|
Serpin family D member 1 |
3.9 |
|
Serpin family A member 3 |
2.7 |
|
Inter-alpha-trypsin inhibitor heavy chain 3 |
3.6 |
|
Inter-alpha-trypsin inhibitor heavy chain 2 |
3.2 |
|
Hemopexin |
3.1 |
|
Apolipoprotein H |
−18.8 |
|
Alpha-2-macroglobulin |
−2.1 |
References
- 1. Tenter AM, Heckeroth AR, Weiss LM.
Toxoplasma gondii: from animals to humans. Int J Parasitol 2000;30:1217-1258.
- 2. Jackson MH, Hutchison WM. The prevalence and source of Toxoplasma infection in the environment. Adv Parasitol 1989;28:55-105.
- 3. Chung WH, Hung SI, Yang JY, Su SC, Huang SP, Wei CY, Chin SW, Chiou CC, Chu SC, Ho HC, Yang CH, Lu CF, Wu JY, Liao YD, Chen YT. Granulysin is a key mediator for disseminated keratinocyte death in Stevens-Johnson syndrome and toxic epidermal necrolysis. Nat Med 2008;14:1343-1350.
- 4. Decoster A, Darcy F, Capron A. Recognition of Toxoplasma gondii excreted and secreted antigens by human sera from acquired and congenital toxoplasmosis: identification of markers of acute and chronic infection. Clin Exp Immunol 1988;73:376-382.
- 5. Darcy F, Deslee D, Santoro F, Charif H, Auriault C, Decoster A, Duquesne V, Capron A. Induction of a protective antibody-dependent response against toxoplasmosis by in vitro excreted/secreted antigens from tachyzoites of Toxoplasma gondii
. Parasite Immunol 1988;10:553-567.
- 6. Duquesne V, Auriault C, Darcy F, Decavel JP, Capron A. Protection of nude rats against Toxoplasma infection by excreted-secreted antigen-specific helper T cells. Infect Immun 1990;58:2120-2126.
- 7. Rahmah N, Anuar AK. Demonstration of antigenic similarities and variations in excretory/secretory antigens of Toxoplasma gondii
. Biochem Biophys Res Commun 1992;187:294-298.
- 8. Zenner L, Estaquier J, Darcy F, Maes P, Capron A, Cesbron-Delauw MF. Protective immunity in the rat model of congenital toxoplasmosis and the potential of excreted-secreted antigens as vaccine components. Parasite Immunol 1999;21:261-272.
- 9. Cesbron-Delauw MF, Capron A. Excreted/secreted antigens of Toxoplasma gondii--their origin and role in the host-parasite interaction. Res Immunol 1993;144:41-44.
- 10. Ossorio PN, Dubremetz JF, Joiner KA. A soluble secretory protein of the intracellular parasite Toxoplasma gondii associates with the parasitophorous vacuole membrane through hydrophobic interactions. J Biol Chem 1994;269:15350-15357.
- 11. Hoppe HC, Ngô HM, Yang M, Joiner KA. Targeting to rhoptry organelles of Toxoplasma gondii involves evolutionarily conserved mechanisms. Nat Cell Biol 2000;2:449-456.
- 12. Prigione I, Facchetti P, Lecordier L, Deslée D, Chiesa S, Cesbron-Delauw MF, Pistoia VJ. T cell clones raised from chronically infected healthy humans by stimulation with Toxoplasma gondii excretory-secretory antigens cross-react with live tachyzoites: characterization of the fine antigenic specificity of the clones and implications for vaccine development. Immunol 2000;164:3741-3748.
- 13. Mercier C, Cesbron-Delauw MF, Sibley LD. The amphipathic alpha helices of the Toxoplasma protein GRA2 mediate post-secretory membrane association. J Cell Sci 1998;111:2171-2180.
- 14. Nockemann S, Dlugonska H, Henrich B, Kitzerow A, Däubener W. Expression, characterization and serological reactivity of a 41 kDa excreted-secreted antigen (ESA) from Toxoplasma gondii
. Mol Biochem Parasitol 1998;97:109-121.
- 15. Lee WK, Ahn HJ, Baek JH, Lee CH, Yu YG, Nam HW. Comprehensive Proteome Analysis of the Excretory/Secretory Proteins of Toxoplasma gondii
. Bull Korean Chem Soc 2014;35:3071-3076.
- 16. Yang Z, Ahn HJ, Park YH, Nam HW. Afatinib reduces STAT6 signaling of host ARPE-19 cells infected with Toxoplasma gondii
. Korean J Parasitol 2016;54:31-38.
- 17. Yang Z, Ahn HJ, Nam HW. Gefitinib inhibits the growth of Toxoplasma gondii in HeLa cells. Korean J Parasitol 2014;52:439-441.
- 18. Kim YH, Bhatt L, Ahn HJ, Yang Z, Lee WK, Nam HW. Suppressors for human epidermal growth factor receptor 2/4 (HER2/4): a new family of anti-toxoplasmic agents in ARPE-19 cells. Korean J Parasitol 2017;55:491-503.
- 19. Song KJ, Ahn HJ, Nam HW. Anti-apoptotic effects of SERPIN B3 and B4 via STAT6 activation in macrophages after infection with Toxoplasma gondii
. Korean J Parasitol 2012;50:1-6.
- 20. Ahn HJ, Kim JY, Ryu KJ, Nam HW. STAT6 activation by Toxoplasma gondii infection induces the expression of Th2 C-C chemokine ligands and B clade serine protease inhibitors in macrophage. Parasitol Res 2009;105:1445-1453.
- 21. Moumen A, Masterson P, O’Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 2005;123:1065-1078.
- 22. Guo Z, Neilson LJ, Zhong H, Murray PS, Zanivan S, Zaidel-Bar R. E-cadherin interactome complexity and robustness resolved by quantitative proteomics. Sci Signal 2014;7:rs7.
- 23. Pasterkamp RJ, Peschon JJ, Spriggs MK, Kolodkin AL. Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature 2003;424:398-405.
- 24. Suzuki K, Okuno T, Yamamoto M, Pasterkamp RJ, Takegahara N, Takamatsu H, Kitao T, Takagi J, Rennert PD, Kolodkin AL, Kumanogoh A, Kikutani H. Semaphorin 7A initiates T-cell-mediated inflammatory responses through alpha1beta1 integrin. Nature 2007;446:680-684.
- 25. Scott GA, McClelland LA, Fricke AF. Semaphorin 7a promotes spreading and dendricity in human melanocytes through beta1-integrins. J Invest Dermatol 2008;128:151-161.
- 26. Isenberg J, Golizeh M, Belfort RN, da Silva AJ, Burnier MN, Ndao M. Peptidyl-prolyl cis-trans isomerase A - A novel biomarker of multi-episodic (recurrent) ocular toxoplasmosis. Exp Eye Res 2018;177:104-111.
- 27. Demarchi F, Bertoli C, Copetti T, Eskelinen EL, Schneider C. Calpain as a novel regulator of autophagosome formation. Autophagy 2007;3:235-237.
- 28. Cataldo F, Peche LY, Klaric E, Brancolini C, Myers MP, Demarchi F, Schneider C. CAPNS1 regulates USP1 stability and maintenance of genome integrity. Mol Cell Biol 2013;33:2485-2496.