Abstract
Echinostoma mekongi n. sp. (Digenea: Echinostomatidae) is described based on adult flukes collected from humans residing along the Mekong River in Cambodia. Total 256 flukes were collected from the diarrheic stool of 6 echinostome egg positive villagers in Kratie and Takeo Province after praziquantel treatment and purging. Adults of the new species were 9.0–13.1 (av. 11.3) mm in length and 1.3–2.5 (1.9) mm in maximum width and characterized by having a head collar armed with 37 collar spines (dorsal spines arranged in 2 alternative rows), including 5 end group spines. The eggs in feces and worm uterus were 98–132 (117) μm long and 62–90 (75) μm wide. These morphological features closely resembled those of Echinostoma revolutum, E. miyagawai, and several other 37-collar-spined Echinostoma species. However, sequencing of the nuclear ITS (ITS1-5.8S rRNA-ITS2) and 2 mitochondrial genes, cox1 and nad1, revealed unique features distinct from E. revolutum and also from other 37-collar-spined Echinostoma group available in GenBank (E. bolschewense, E. caproni, E. cinetorchis, E. deserticum, E. miyagawai, E. nasincovae, E. novaezealandense, E. paraensei, E. paraulum, E. robustum, E. trivolvis, and Echinostoma sp. IG). Thus, we assigned our flukes as a new species, E. mekongi. The new species revealed marked variation in the morphology of testes (globular or lobulated), and smaller head collar, collar spines, oral and ventral suckers, and cirrus sac compared to E. revolutum and E. miyagawai. Epidemiological studies regarding the geographical distribution and its life history, including the source of human infections, remain to be performed.
-
Key words: Echinostoma mekongi, 37-collar-spined echinostome, Kratie Province, Takeo Province, Mekong River, Cambodia
INTRODUCTION
Echinostoma spp. (family Echinostomatidae) of the 37-collar-spined group, or ‘
Echinostoma revolutum group’, are taxonomically diverse consisting of at least 15 (excluding
E. mekongi of this study) valid and 10 validity-retained species worldwide [
1]. The 15 valid species include
E. revolutum (Froelich, 1802) Dietz, 1909,
E. bolschewense (Kotova, 1939) Nasincova, 1991,
E. caproni Richard, 1964,
E. cinetorchis Ando & Ozaki, 1924,
E. deserticum Kechemir et al., 2002,
E. lindoense Sandground & Bonne, 1940,
E. luisreyi Maldonado et al., 2003,
E. miyagawai Ishii, 1932,
E. nasincovae Faltýnková et al., 2015,
E. novaezealandense Georgieva et al., 2017,
E. paraensei Lie & Basch, 1967,
E. paraulum Dietz, 1909,
E. robustum Yamaguti, 1935,
E. trivolvis (Cort, 1914) Kanev, 1985, and
Echinostoma sp. IG of Georgieva et al., 2013 [
1]. All of these species, except
E. lindoense and
E. luisreyi, have unique molecular data deposited in GenBank. The 10 validity-retained species include
E. acuticauda Nicoll, 1914,
E. barbosai Lie & Basch, 1966,
E. chloephagae Sutton & Lunaschi, 1980,
E. echinatum (Zeder, 1803) de Blainville, 1828,
E. jurini (Skvortsov, 1924) Kanev, 1985,
E. nudicaudatum Nasir, 1960,
E. parvocirrus Nassi & Dupouy, 1988,
E. pinnicaudatum Nasir, 1961,
E. ralli Yamaguti, 1934, and
E. rodriguesi Hsu et al., 1968. Seven species among them, including
E. revolutum,
E. cinetorchis,
E. echinatum (needs confirmation),
E. lindoense,
E. miyagawai (experimental infection),
E. paraulum, and possibly
E. paraensei (from the coprolite of a human mummy), were regarded as human-infecting zoonotic echinostomes [
1–
3].
The species differentiation of 37-collar-spined echinostomes has been done mostly based on the morphology, biology, life-cycle, and host characteristics of each species [
4]. However, identification of these species only by morphology is difficult and confusing, and molecular analyses are a highly useful alternative method [
5,
6]. When sequences of the nuclear internal transcribed spacer region (ITS1-5.8S rRNA-ITS2) or mitochondrial genes, including cytochrome
c oxidase 1 (
cox1) and NADH dehydrogenase subunit 1 (
nad1), were analyzed, remarkable genetic variation has been noted in
E. revolutum and other 37-collar-spined
Echinostoma spp. from Europe [
7–
9], North America [
10,
11], Oceania [
6,
12], Southeast Asia [
5,
13–
15], and various continents and localities [
16]. There were 2 genetic lineages of
E. revolutum; Eurasian and American by
nad1 and Southeast Asian and American by
cox1 analyses [
5].
E. miyagawai also revealed 2 genetic lineages, Australian and Eurasian by
nad1 analysis [
5]. Other 37-collar-spined group deposited in GenBank, which are genetically distinct, include
E. bolschewense,
E. caproni,
E. cinetorchis,
E. deserticum,
E. nasincovae,
E. novaezealandense,
E. paraensei,
E. paraulum,
E. robustum,
E. trivolvis, and
Echinostoma IG of Georgieva et al., 2013 [
1,
5–
12].
We collected adult flukes of 37-collar-spined Echinostoma species from 6 riparian people in 2 localities (Kratie and Takeo Province) of Cambodia along the Mekong River, after treatment with praziquantel and purging. The flukes were morphologically difficult to differentiate from E. revolutum and several other members of 37-collar-spined Echinostoma group. Therefore, the nuclear ITS gene and 2 mitochondrial genes (cox1 and nad1) were sequenced, and the flukes were found to be genetically distinct from the 13 ever-known 37-collar-spined Echinostoma species available in GenBank, including E. revolutum and E. miyagawai. The other species of 37-collar-spined Echinostoma group unavailable in GenBank were morphologically differed from our flukes. Therefore, our flukes were assigned as a new species, i.e., E. mekongi n. sp.
MATERIALS AND METHODS
Patients and worm recovery
A total of 256 adult flukes of 37-collar-spined echinostomes,
E. mekongi n. sp., were collected from 6 riparian people (1–157 worms by individual) living along the Mekong River (4 people from Kratie Province and 2 from Takeo Province), Cambodia in May 2011 (
Table 1). Some of them complained of abdominal discomfort, indigestion, and other mild gastrointestinal troubles; the others had little clinical symptoms. The procedures of worm recovery were as described previously [
17]. Some flukes were fixed in 10% neutral formalin for morphological studies, and the others were fixed and preserved in 70–80% ethanol for molecular analyses. The formalin-fixed samples were washed with water and stained with Semichon’s acetocarmine, dehydrated with a graded series of ethanol, cleared in xylene, and mounted in Permount. The ethanol-fixed samples were used for molecular analyses. Informed consent was obtained from each enrolled person or guardians. The procedures of worm collection from the people were permitted under the agreement between the National Center for Parasitology, Entomology, and Malaria Control, Phnom Penh, Cambodia and the Korea Association of Health Promotion, Seoul, the Republic of Korea (2009–2011).
Twenty-six acetocarmine-stained specimens were used for morphological observations, measurements (
Table 2), and description of worms. The comparison of our specimens with 37-collar-spined
Echinostoma spp. was based on morphological characters given by previous authors [
4,
6,
18]. Photomicrographs of the worms were taken with a digital camera (Olympus DP72, Tokyo, Japan) on an Olympus CKX41 microscope (Tokyo, Japan). Measurements were taken from digital images with the aid of CellSens Standard v1.5 image analysis software.
The following morphological characters and measurements were used for species comparison of 37-collar-spined
Echinostoma group [
6,
18]. They included the body length, body width at 3 different levels (intestinal bifurcation, posterior border of ventral sucker, and mid-way between ventral sucker and ovary), head collar, angle spine, lateral spine, dorsal spine, oral sucker, pre-pharynx length, pharynx, esophagus length, cirrus-sac, seminal vesicle, ventral sucker, ovary, Mehlis’ gland, anterior testis, posterior testis, eggs, forebody length, distance from anterior extremity to intestinal bifurcation, distance between posterior margin of ventral sucker and ovary, length of post-testicular region, oral sucker to pharynx width ratio, body width to body length ratio, forebody length to body length, collar width to maximum body width ratio, distance from anterior extremity to intestinal bifurcation to body length ratio, length of pre-ovarian region to body length ratio, and length of post-testicular region to body length ratio [
18].
For molecular analyses, worms preserved in 70–80% ethanol were used. If combined morphological and molecular analyses were preferred, worms mounted on glass slides were photographed and then removed from the slides using a slide heater or xylene for molecular analyses. Genomic DNA was extracted using the Spin-Column Protocol of DNeasy® Blood & Tissue kit (QIAGEN, Hilden, Germany). PCR and nested-PCR were then conducted using specific primers designed to amplify ITS (ITS1-5.8S rRNA-ITS2) [
19] and
cox1 and
nad1 [
16] genes in echinostomes. The primers for ITS gene were BD1 and BD2 [
19], and the those for
cox1 were JB3 and JB13 [
16]. The primers for
nad1 were JB11 and JB12 in the first PCR and EchND1/inF and EchND1/inR for the second PCR of the inner region [
16]. The PCR products were sequenced using the BigDye® Terminator v3.1 cycle sequencing kit by ABI 3730XL DNA analyzer (Applied Biosystems, Foster City, California, USA). For evaluation of the genetic identity of the samples, the basic local alignment search tool (BLAST;
http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used. Using the Geneious® version 6.1.6 (Biometers Ltd., Auckland, New Zealand), we aligned the obtained sequences with GenBank reference ITS,
cox1, and
nad1 sequences of 37-collar-spined
Echinostoma species. Phylogenetic information was assessed via maximum-likelihood (ML) analyses using the MEGA v6 program applying Tamura-nei model of nucleotide substitution with 1,000 bootstrap replications [
20].
RESULTS
Description of worms
Family Echinostomatidae Looss, 1899
Subfamily Echinostomatinae Looss, 1899
Genus Echinostoma Rudolphi, 1809
Adults
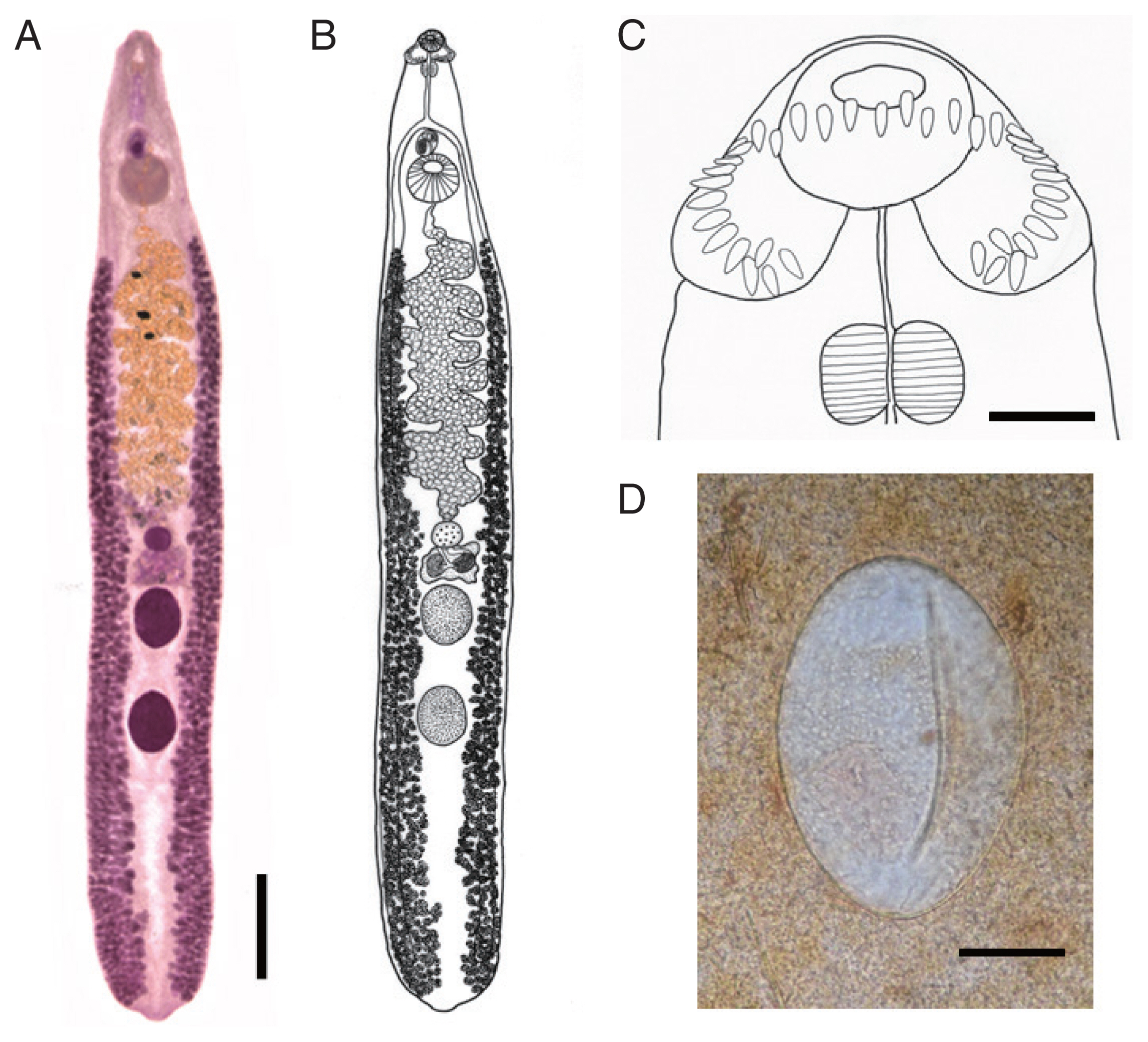
Body dorsoventrally flattened, muscular, elongated leaf-like with slightly attenuated both ends, 8.97–13.12 (av. 11.28) mm in length and 1.34–2.50 (1.93) mm in maximum width at mid-uterine or ovarian region (n=20) (
Table 2;
Figs. 1A, B,
2). Tegument beset with triangular spines, less dense posteriorly, extending to level of posterior testis. Forebody long representing about 17.5% of whole body length. Anterior end with characteristic features of an echinostome, equipped with an oral sucker and a head collar. Oral sucker small, muscular, spherical, subterminal, about 2/5 of the size of the ventral sucker. Head collar small but prominent, muscular, and reniform armed with collar spines (
Fig. 1C). Collar spines 37 in total number, with the formula of 5-6-6-3-6-6-5, including angle (corner) spines 5+5, lateral spines 6+6, dorsal spines 6+3+6; dorsal spines arranged in 2 alternating rows (
Fig. 1C). Collar spines relatively small in length and width, moderately pointed (not sharply pointed nor blunt-ended). Prepharynx relatively long; pharynx muscular, elongated oval. Esophagus long; intestinal bifurcation from anterior extremity at about 13% of total body length; ceca blind, narrow, overlapped by vitelline follicles, ending before the posterior termination of vitelline follicles. Cirrus sac transversely oval, with muscular walls, located between intestinal bifurcation and anterior margin of ventral sucker, containing seminal vesicle (with elongate-oval anterior portion and narrow saccular posterior portion), well-developed pars prostatica, coiled ejaculatory duct, muscular cirrus with smooth unarmed surface. Genital atrium and genital pore median, just posterior to intestinal bifurcation, receiving female (metraterm) and male reproductive (ejaculatory) duct. Uterus intercecal, long, slender, with numerous transverse coils between ventral sucker and ovary, containing a large number of eggs. Metraterm short, weakly muscular, connected to genital pore. Ovary spherical, median or slightly submedian, almost equatorial, between uterus and Mehlis’ gland. Mehlis’ gland transversely oval, median, connected with ovary and uterine tubule. Uterine seminal receptacle present, ventral to Mehlis’ gland; Laurer’s canal absent. Vitelline follicles extensive, extending laterally forming 2 lateral groups, from the level slightly posteriorly to ventral sucker to near posterior extremity; the 2 groups of vitellaria not merge until their posterior extremities. Two testes tandem, globular, entire in more than 2/3 of the specimens or 5–8 lobulated in about 1/3 of the specimens, located in posterior field of body, more or less separated from each other. Eggs numerous, yellowish, immature containing a germ cell, operculate with a small, inconspicuous operculum, and a small abopercular thickening or wrinkling at the abopercular end, oval, 98–132 (117) μm long and 62–90 (75) μm wide (n=20) (
Fig. 1D). Excretory vesicle Y-shaped, bifurcates just posterior to posterior testis; excretory pore terminal.
Type host: Homo sapiens (natural infection)
Site of infection: Small intestine
Type locality: Kratie and Takeo Province, Cambodia
Deposition of specimens: The type specimen is deposited in the Parasite Museum, Institute of Parasitic Diseases, Korea Association of Health Promotion, Seoul, Republic of Korea (no. 2020-0011-01, holotype, and no. 2020-0011-02~24, subtypes). Voucher specimens are deposited in Meguro Parasitological Museum, Tokyo, Japan (MPM Coll. No. 21675).
Etymology: The specific name refers to the name of the river (the Mekong River); the infected humans were residents living nearby the river.
Molecular analyses
A phylogenetic tree based on ITS region (992 bp after trimming) was constructed using the ML method. The tree comprised of sequences of 7 species of 37-collar-spined
Echinostoma group, including our samples (
E. mekongi n. sp.). The sequences of our samples (10 isolates; GenBank accession nos. MT409010-409019) clustered together (100% homologous) with high bootstrap values and constructed a new lineage distinct from any other 37-collar-spined
Echinostoma species deposited in GenBank (
Fig. 3). Although
E. mekongi n. sp. appeared to be a sister group to
E. revolutum, a morphologically highly similar species, the sequence comparison showed only 97.6% similarity between the 2 species (
Table 3). In addition, the sequences of
E. paraensei and
E. robustum showed 96.4% and 97.9% similarities with the new species, respectively (
Table 3).
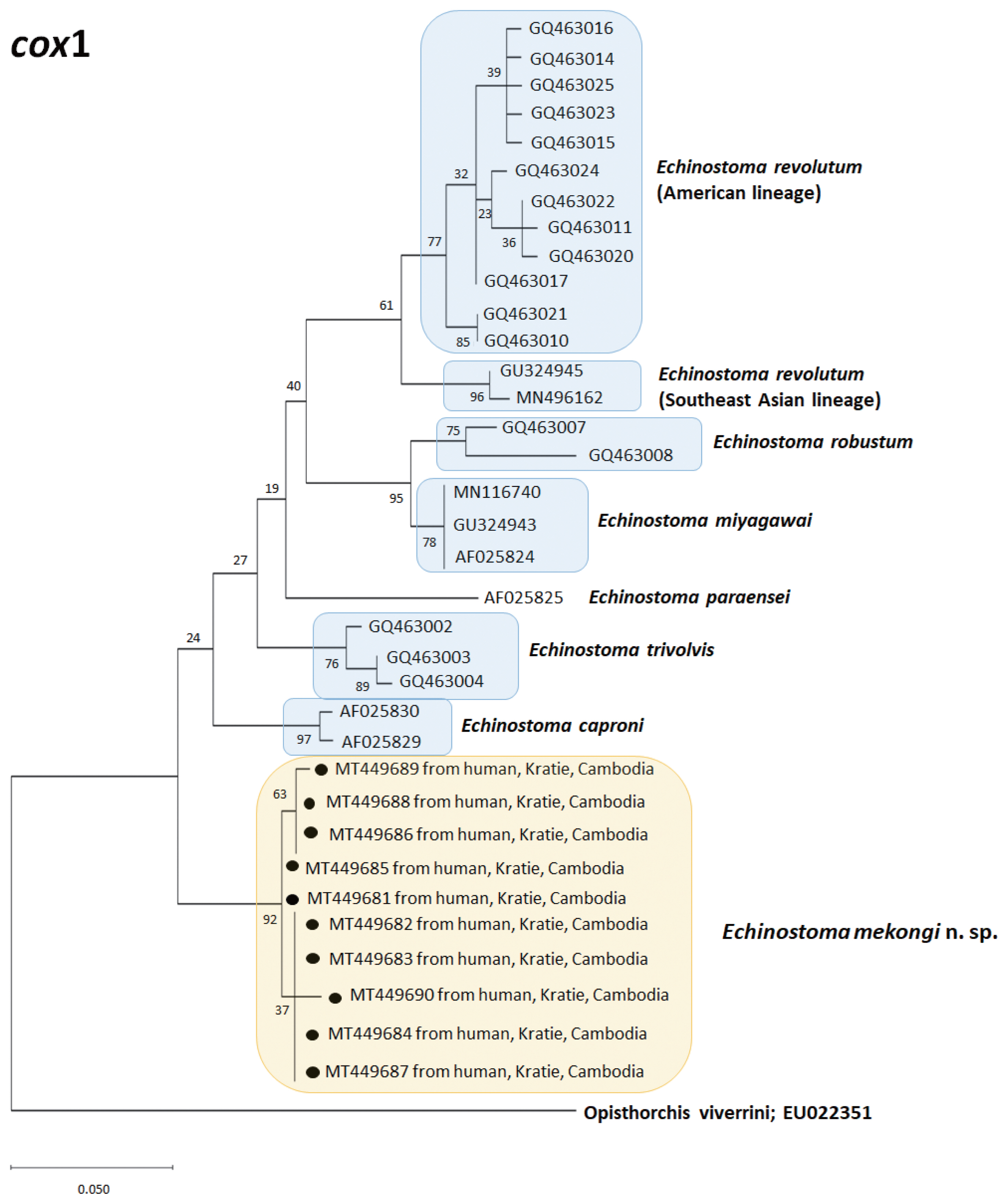
A phylogenetic tree based on
cox1 (184 bp after trimming) sequences was constructed using the ML analyses. The tree comprised of 7 species of 37-collar-spined
Echinostoma group, including Southeast Asian and American lineages of
E. revolutum and
E. mekongi n. sp. The sequences of our samples (10 isolates; GenBank accession nos. MT449681-449690) clustered together (99.6%) with high bootstrap values and constructed a new lineage distinct from any other 37-collar-spined
Echinostoma species deposited in GenBank (
Fig. 4). Although our samples appeared to be a sister group to
E. caproni, the sequence showed only 92.6–92.7% similarity between the 2 species (
Table 3). In addition, the sequences of
E. trivolvis,
E. paraensei,
E. miyagawai,
E. robustum, and
E. revolutum having similar morphology with our samples showed only 89.3–92.7% homology with our samples (
Table 3).
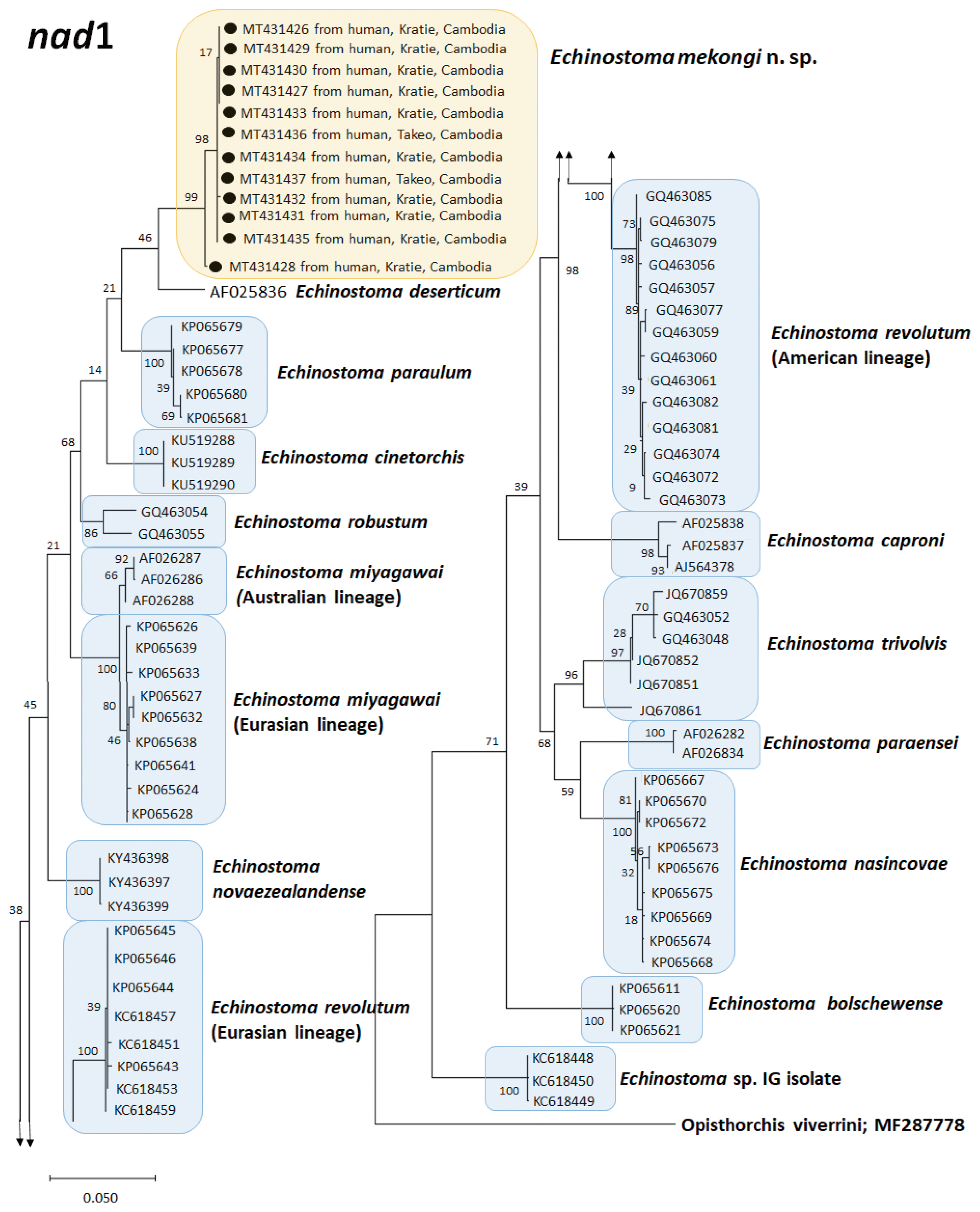
A phylogenetic tree based on
nad1 (472 bp after trimming) sequences constructed using the ML method is shown in
Fig. 5. The tree composed of sequences of 14 different species of 37-collar-spined
Echinostoma group available in GenBank, including
E. mekongi n. sp., Eurasian and Australian lineages of
E. miyagawai, and Eurasian and American lineages of
E. revolutum. The sequences of our samples (12 isolates; GenBank accession nos. MT431426-431437) clustered together (99.4–99.8%) with high bootstrap values constructing a new genetic lineage distinct from any other 37-collar-spined
Echinostoma species (
Fig. 5). In this
nad1 phylogenetic tree, the lineage of our samples appeared as a sister group to
E. deserticum; however, the sequence similarity between the 2 was only 87.1–87.2% (
Table 3). In addition, the sequences of
E. revolutum and
E. miyagawai having similar morphologies with our samples showed only 85.7–86.6% and 86.0–87.7% sequence homologies with our samples, respectively (
Table 3).
Compared with the 15 valid and 10 validity-retained species of ‘
E. revolutum group’, the new species morphologically differed from most of them, in particular, those reported from Europe, Africa, North and South America, and Oceania [
1]. However, the new species appeared to be comparable with those reported from Asia which included
E. revolutum,
E. cinetorchis,
E. lindoense,
E. miyagawai,
E. paraulum,
E. ralli, and
E. robustum [
1]. The new species most closely resembled 4 of them,
E. revolutum,
E. lindoense,
E. miyagawai, and
E. paraulum (
Table 2).
E. cinetorchis differed from
E. mekongi in that
E. cinetorchis has abnormally located or reduced number of testes [
1–
3] whereas no such specimen was found in the new species.
E. ralli differed from
E. mekongi in that the former has 4 end group spines [
21], whereas the new species has 5 end group spines.
E. robustum could be distinguished from
E. mekongi in the morphology of testes, globular or slightly lobulated, and vitellaria not merging post-testicularly in the new species and irregularly lobed and horizontally extended testes and 2 lateral vitellaria merging beyond the posterior testis level in
E. robustum [
22].
E. mekongi differed from the 4 closely related species in that it had a smaller head collar (av. 555 μm in collar width) compared to
E. revolutum (av. 607 μm),
E. miyagawai (av. 656 μm),
E. paraulum (av. 729 μm), and
E. lindoense (780 μm), and smaller collar spines in comparison with the same 4 species (
Table 2). The collar spines of
E. mekongi were not so long and not sharply pointed as those of
E. revolutum or
E. miyagawai [
1]. The oral and ventral suckers were also smaller (av. 244×231 μm and 603×573 μm, respectively) compared with those of
E. revolutum (av. 316×282 μm and 893×873 μm, respectively),
E. miyagawai (av. 305×293 μm and 738×706 μm, respectively), and
E. paraulum (av. 335×287 μm and 731×667 μm, respectively) [
18]. The cirrus sac was also smaller (av. 401×249 μm) than that of
E. revolutum (av. 552×316 μm),
E. miyagawai (av. 506×300 μm), and
E. paraulum (av. 413×281 μm) [
18]. The egg size of
E. mekongi (av. 117×75 μm) was similar to the eggs of
E. revolutum,
E. paraulum, and
E. lindoense but larger than that of
E. miyagawai (av. 95×60 μm) [
18,
23] (
Table 2). The forebody length, the distance from the anterior extremity to intestinal bifurcation, and the distance from the posterior margin of ventral sucker to ovary were longer in
E. mekongi than in
E. revolutum,
E. miyagawai, and
E. paraulum but almost equal to or shorter than that in
E. lindoense [
18,
23] (
Table 2). The vitelline follicles of
E. mekongi were distributed laterally from the level of some distance from the posterior margin of the ventral sucker to the posterior extremity, without merging beyond the posterior testis level which resembled
E. revolutum and
E. paraulum [
1,
18]. By comparison, the 2 lateral groups of vitelline follicles in
E. miyagawai and
E. lindoense merge near the posterior extremity [
1].
Three phylogenetic trees based on ITS region,
cox1, and
nad1 sequences revealed a unique genetic lineage of
E. mekongi distinguished from all other species of 37-collar-spined
Echinostoma spp. (
Figs. 3–
5). Even in ITS region in which interspecific variation is not so remarkable, the sequence homologies between
E. mekongi and other species such as
E. revolutum,
E. robustum,
E. caproni,
E. trivolvis, and
E. paraensei were lower than 97.9% (
Table 3). The
cox1 sequences of
E. mekongi revealed lower than 92.7% homologies in comparison with
E. revolutum (Southeast Asian and American lineages; Nagataki et al. [
5]) and 5 other species, and the
nad1 sequences showed lower than 88.2% homologies compared with
E. revolutum (Eurasian and American lineages; Nagataki et al. [
5]),
E. miyagawai (Eurasian and Australian lineages; Nagataki et al. [
5]), and 11 other species deposited in GenBank. Therefore,
E. mekongi is considered a new species morphologically and molecularly distinct from the pre-existing 37-collar-spined
Echinostoma spp. reported around the world.
DISCUSSION
Kanev [
4] reported that the species of ‘
E. revolutum group’ cannot be morphologically identified only by adult flukes but can be more clearly discriminated by the morphology of larval forms, in particular, the cercariae, and host-parasite relationships. In the cercariae, the numbers of penetration gland-cell outlets and paraesophageal gland-cell outlets, flame cell patterns, and the presence or absence as well as the number of tail fin-folds are important criteria for species differentiation [
4,
24]. However, an integrated taxonomic approach, linking morphological and molecular evidence, was also important in assessing the species diversity within the ‘
E. revolutum’ group from Europe [
18]. In the present study, this integrated approach was extremely useful for establishing a new species (
E. mekongi) from Asia, particularly in the absence of larval stage information, including the larval morphology and intermediate hosts.
The new species was found from riparian people along the Mekong River. Most of the patients were young females at the age of the 10s-20s, except one who was a 37-year-old female. They recalled that they had eaten various kinds of snails (species unknown) purchased from local markets which were stated to have been caught around the river. Snails (gastropods and bivalves) have been reported to be the first and/or second intermediate host for most of the ‘
E. revolutum group’ [
1,
4]. However, the larval stages, including the rediae, cercariae, and metacercariae, of
E. mekongi have not yet been discovered, and the source of infection in our patients remains to be determined. The patients complained of abdominal discomfort, indigestion, and other mild gastrointestinal symptoms, although there is no evidence whether these symptoms were directly related to
E. mekongi infection or not. However, it is referable that in
Isthmiophora hortensis (under the name
Echinostoma hortense) infection, the patients complained of severe epigastric discomfort with ulcerative lesions in the stomach or the duodenum, and living worms were detected near the ulcerative lesions through gastroduodenal endoscopy [
2,
3]. Moreover, in
Artyfechinostomum malayanum (under the name
Artyfechinostomum mehrai) infection, a fatal human case was reported in India in whom marked malnutrition and anemia were found and several hundred worms were collected at autopsy [
25].
With designation of our flukes as a new species of ‘
E. revolutum group’, it is interesting to consider its possible wider geographical distribution from Kratie and Takeo Province to other provinces of Cambodia as well as to other Indochina countries, including Vietnam, Lao PDR, and Thailand. In addition, the specific diagnosis of 37-collar-spined echinostomes reported previously from Indochina peninsula needs reconfirmation. For example, in Cambodia, human infections with
E. ilocanum were reported among inhabitants in Oddar Meanchey Province [
26] and human infections with
E. revolutum were reported among schoolchildren in Pursat Province [
27]. The morphological diagnosis of
E. ilocanum seems to be of no problem because the worms characteristically had 49–51 collar spines [
26]. However, the diagnosis of
E. revolutum was based only on worm morphology [
27] and needs further verification using molecular analyses.
In Vietnam, at least 3 papers reported the presence of 37-collar-spined
Echinostoma spp. [
28–
30]. Two were reports of
E. cinetorchis infection in dogs in Nghe An Province [
28] and in chickens and ducks in Nam Dinh Province [
29]. The first paper shows a figure of
E. cinetorchis [
28] but based on the figure we consider that the diagnosis had not been properly given for them. The second paper does not show any figure of
E. cinetorchis nor any description of the worms [
29]; thus, we cannot assure the specific diagnosis. The third paper was on the partial life cycle of
E. revolutum, with the metacercariae found in
Filopaludina snails in Nam Dinh Province and adults obtained from experimentally infected hamsters [
30]. Because the adult worms are morphologically difficult to distinguish from those of
E. mekongi, further confirmation of the species using molecular methods seems to be needed.
In Lao PDR, very few papers were published regarding the existence of ‘
E. revolutum group’. One was the report of human
E. revolutum infections who were co-infected with
A. malayanum and others [
17]. Other echinostome species reported from Lao PDR included
Echinochasmus japonicus [
31],
Echinostoma macrorchis [
32],
Echinostoma ilocanum [
33], E
chinostoma aegyptica [
34], and
Echinochasmus caninus [
35]. The diagnosis of all these echinostomes was based on the morphology of adult flukes. Among them, the diagnosis of
E. revolutum [
17] remains to be reconfirmed through molecular studies.
In Thailand, many papers have reported human and animal (ducks) infections with
E. revolutum [
2,
5,
13,
14,
36–
39]. Among them, molecular data were provided by Saijuntha and co-workers [
13,
38], Noikong et al. [
14], Nagataki et al. [
5], and Buddhachat and Chontananarth [
39]. Regarding the isolate of
E. revolutum by Saijuntha et al. [
38] which exhibited close affinity to the European isolate studied by Morgan and Blair [
12,
16], Georgieva et al. [
9] pointed out that it was shown to represent
E. miyagawai rather than
E. revolutum. In addition, the molecular data presented by Noikong et al. [
14] were not deposited in GenBank, so there is no evidence to confirm the diagnosis of
E. trivolvis/
revolutum-like clade among their samples [
9]. However, Nagataki et al. [
5] demonstrated the presence of 2 species of ‘
E. revolutum group’ in Thailand and Lao PDR, which included
E. revolutum and
E. miyagawai. Moreover, from the phylogenetic tree they established, 2 separate genetic lineages of
E. revolutum were found, namely, the Southeast Asian and American lineages by analysis of
cox1 or the Eurasian and American lineages based on
nad1 sequences. Using DNA barcoding conjugated with high-resolution melting analysis, Buddhachat and Chontananarth [
39] further classified
E. revolutum genetic lineages into 3 based on
nad1 sequences, i.e., the Asian, Eurasian, and American lineages. However, the existence of
E. mekongi in Thailand and Lao PDR remains to be determined.
In conclusion, we discovered a new species of ‘E. revolutum group’ from human infections in Kratie and Takeo provinces, Cambodia, i.e., E. mekongi n. sp. This is a new echinostome fauna in Cambodia and the first report of a human-infecting Echinostoma sp. diagnosed by molecular analyses in Southeast Asia. Several previous reports on the existence of E. revolutum in Indochina countries need re-validation through molecular analyses.
Notes
-
CONFLICT OF INTEREST
We have no conflict of interest related to this study.
ACKNOWLEDGMENTS
We are grateful to the staff of National Center for Parasitology, Entomology and Malaria Control, Phnom Penh, Cambodia, and Provincial Health Department, Kratie and Takeo Province, Cambodia who helped this study. We also thank the staff of Korea Association of Health Promotion, Seoul, Korea who performed fecal examinations of people in Kratie and Takeo Province, Cambodia. We also appreciate Hyejoo Shin, Institute of Parasitic Diseases, Korea Association of Health Promotion, Seoul, Korea for her help in line drawing of worm figures.
Fig. 1
Echinostoma mekongi n. sp. from Cambodia. (A) An adult worm (type) collected from a riparian person (37-year-old female). Acetocarmine-stained. Ventral view. Scale bar=1.4 mm. (B) Line drawing of the worm in Fig. 1A. (C) Head collar with 37 collar spines. Scale bar=140 μm. (D) An egg in Kato-Katz fecal smear of a patient from Kratie Province. Scale bar=33 μm.
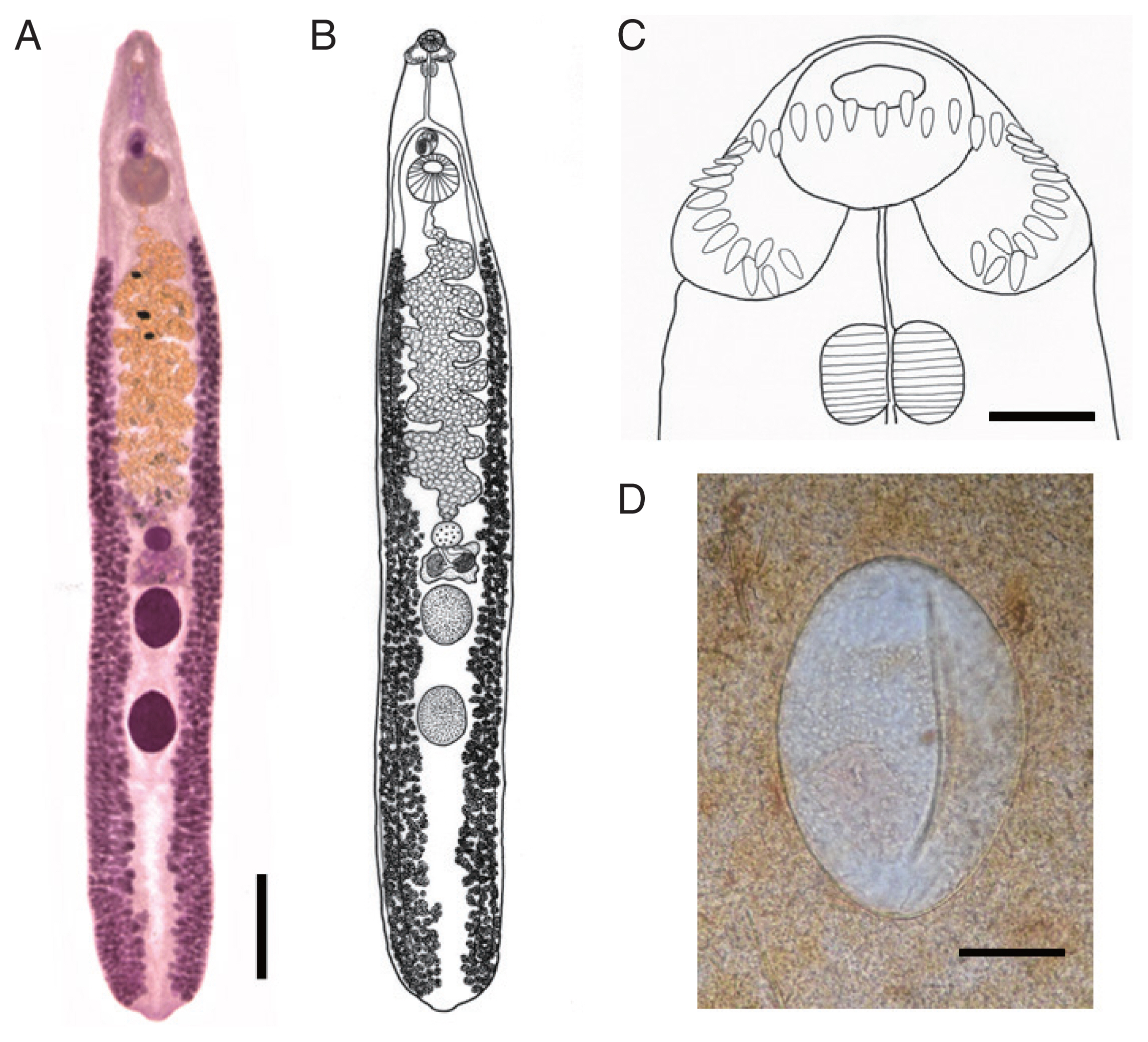
Fig. 2
Echinostoma mekongi n. sp. adult flukes (n=157) collected from a riparian person (37-year-old female) along the Mekong River in Kratie Province, Cambodia.

Fig. 3A phylogenetic tree of Echinostoma mekongi n. sp. and other 37-collar-spined ‘E. revolutum group’ constructed based on 992 bp of nuclear ribosomal ITS (ITS1-5.8S rRNA-ITS2) gene sequences.

Fig. 4A phylogenetic tree of Echinostoma mekongi n. sp. and other 37-collar-spined ‘E. revolutum group’ constructed based on 184 bp of mitochondrial cox1 sequences.
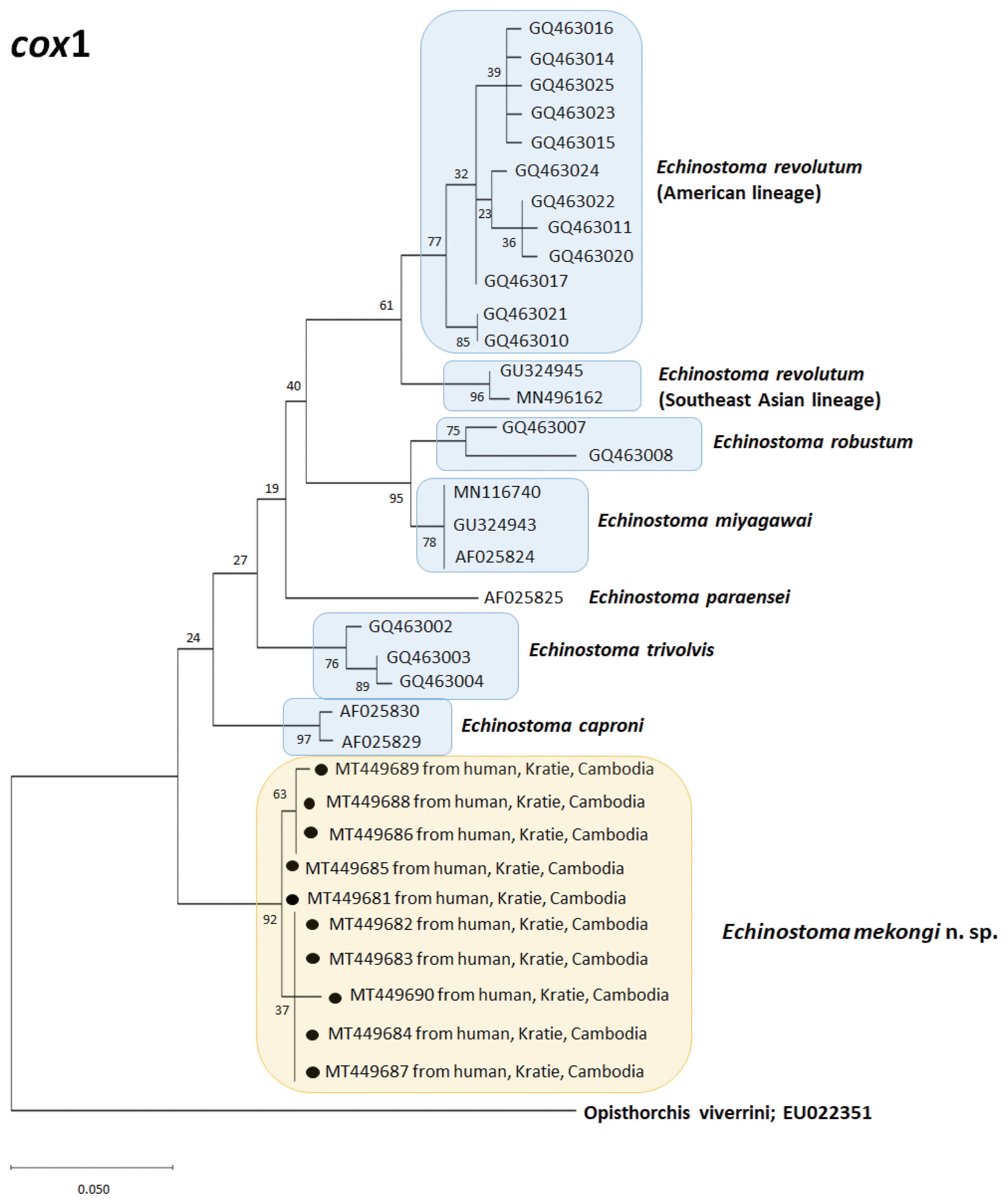
Fig. 5A phylogenetic tree of Echinostoma mekongi n. sp. and other 37-collar-spined ‘E. revolutum group’ constructed based on 472 bp of mitochondrial nad1 sequences.
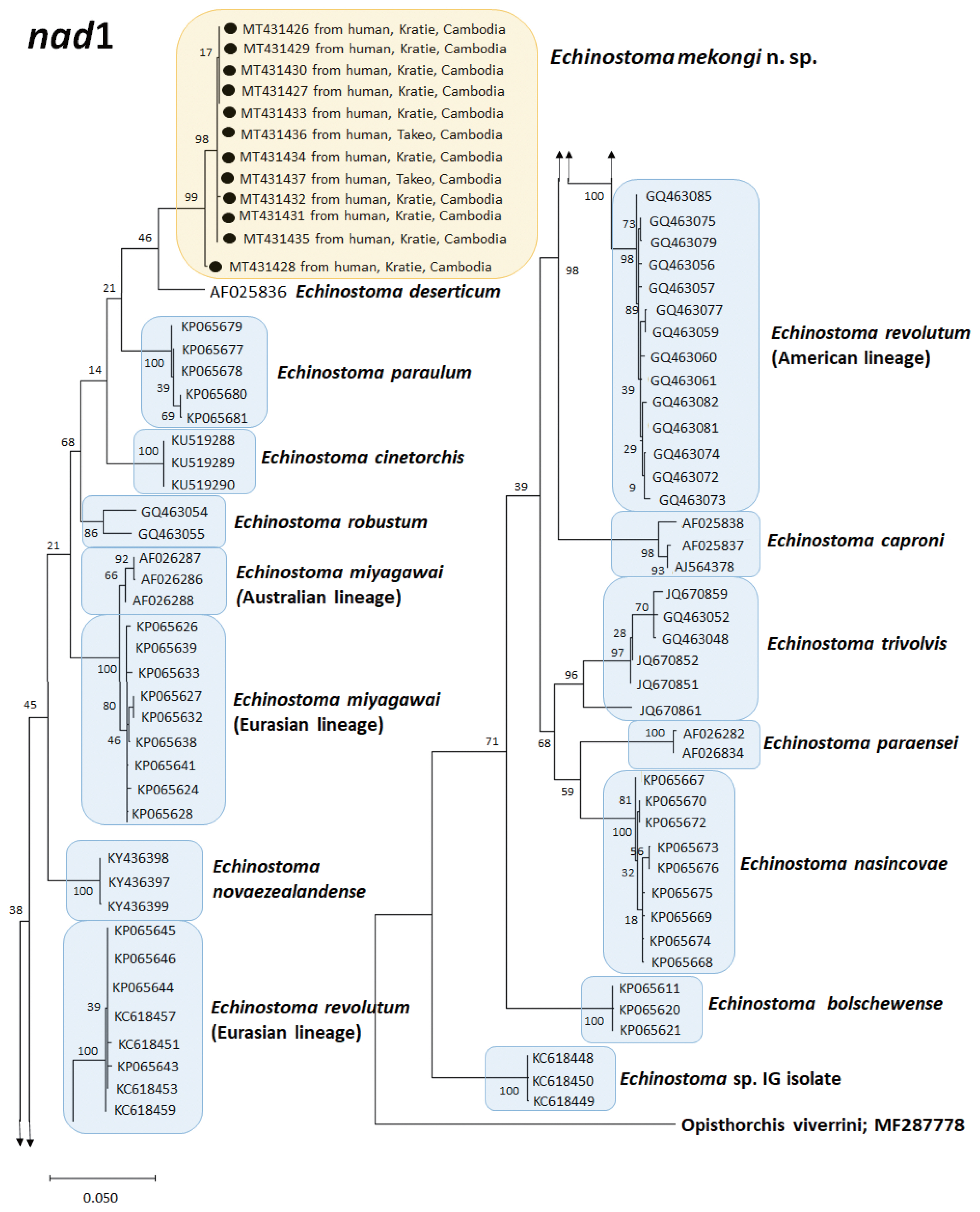
Table 1Worm recovery of Echinostoma mekongi n. sp. from 6 riparian people in 2 provinces of Cambodia
Table 1
|
Province |
Village name |
Patient (age, sex) |
No. of echinostome eggs/gram of fecesa
|
No. of E. mekongi adult specimens collectedb
|
|
Kratie |
Talous |
25 F |
2,976 |
157 |
|
Rokakandal |
37 F |
264 |
53 |
|
Talous |
17 F |
1,896 |
33 |
|
Talous |
15 F |
0 |
1 |
|
|
Takeo |
Ang Svay Chek |
11 F |
1,152 |
6 |
|
Ang Svay Chek |
10 F |
1,032 |
6 |
|
|
Total |
|
|
|
256 |
Table 2Measurements of Echinostoma mekongi n. sp. (adults) in comparison with other 37-collar-spined Echinostoma species (unit: μm)
Table 2
|
Species |
E. mekongi n. sp. |
E. revolutum [18] |
E. miyagawai [18] |
E. paraulum [18] |
E. lindoense [23] |
|
|
|
|
|
|
|
No. of specimens |
n=20 |
n=16 |
n=13 |
n=10 |
Several hundreds |
|
|
|
|
|
|
Mean |
Range |
Mean |
Range |
Mean |
Range |
Mean |
Range |
Range |
|
Body length (BL) |
11,278 |
8,970–13,123 |
10,531 |
9,454–11,846 |
9,990 |
9,163–11,014 |
6,345 |
5,600–6,862 |
13,000–15,000 |
|
|
Body width 1 (BW1) |
1,031 |
774–1,293 |
1,298 |
1,182–1,454 |
1,159 |
1,029–1,272 |
1,043 |
923–1,108 |
2,033a
|
|
|
Body width 2 (BW2) |
1,280 |
903–1,615 |
1,618 |
1,303–1,815 |
1,026 |
898–1,178 |
1,308 |
1,138–1,446 |
2,033a
|
|
|
Body width 3 (BW3) |
1,933 |
1,336–2,503 |
1,815 |
1,454–2,092 |
1,305 |
1,197–1,459 |
1,514 |
1,231–1,662 |
2,500–3,000 |
|
|
Head collar length (CL) |
372 |
336–425 |
392 |
351–424 |
434 |
393–505 |
488 |
432–553 |
390a
|
|
|
Head collar width (CW) |
555 |
468–649 |
607 |
552–652 |
656 |
598–692 |
729 |
644–781 |
780 |
|
|
Oral sucker length (OSL) |
231 |
199–263 |
316 |
261–358 |
305 |
262–337 |
287 |
265–318 |
230–510 (diam.) |
|
|
Oral sucker width (OSW) |
244 |
200–297 |
282 |
246–303 |
293 |
262–355 |
335 |
280–371 |
230–510 (diam.) |
|
|
Angle spine length (ASL) |
54 |
35–86 |
74 |
58–92 |
70 |
52–93 |
100 |
73–124 |
60a
|
|
|
Angle spine with (ASW) |
17 |
9–32 |
17 |
11–24 |
22 |
17–29 |
21 |
15–28 |
18a
|
|
|
Lateral spine length (LSL) |
55 |
40–85 |
79 |
64–91 |
76 |
67–87 |
101 |
85–120 |
62a
|
|
|
Lateral spine width (LSW) |
15 |
9–38 |
17 |
12–21 |
23 |
20–26 |
21 |
17–25 |
20a
|
|
|
Dorsal spine length (DSL) |
45 |
24–95 |
85 |
71–94 |
68 |
58–75 |
99 |
90–105 |
60a
|
|
|
Dorsal spine width (DSW) |
12 |
6–25 |
18 |
13–23 |
21 |
19–26 |
21 |
19–23 |
18a
|
|
|
Prepharynx length (PL) |
119 |
31–149 |
41 |
7–99 |
10 |
0–37 |
45 |
15–68 |
78a
|
|
|
Pharynx length (PHL) |
201 |
170–230 |
237 |
209–258 |
299 |
262–337 |
324 |
273–379 |
180–400 (diam) |
|
|
Pharynx width (PHW) |
179 |
144–217 |
219 |
194–258 |
250 |
187–355 |
238 |
212–280 |
180–400 (diam.) |
|
|
Esophagus length (ESL) |
586 |
333–783 |
408 |
318–569 |
585 |
542–655 |
408 |
265–546 |
519a
|
|
|
Cirrus sac length (CSL) |
401 |
262–564 |
552 |
410–735 |
506 |
468–598 |
413 |
318–531 |
300–500 |
|
|
Cirrus sac width (CSW) |
249 |
190–326 |
316 |
243–403 |
300 |
281–355 |
281 |
205–326 |
300a
|
|
|
Seminal vesicle length (SVL) |
308 |
201–457 |
336 |
224–440 |
- |
- |
238 |
152–303 |
- |
|
|
Seminal vesicle width (SVW) |
147 |
100–216 |
146 |
60–212 |
- |
- |
134 |
114–144 |
- |
|
|
Ventral sucker length (VSL) |
603 |
516–694 |
873 |
796–1,038 |
706 |
655–748 |
667 |
569–766 |
600–1,380 (diam.) |
|
|
Ventral sucker width (VSW) |
573 |
448–648 |
893 |
796–1,061 |
738 |
655–785 |
731 |
705–766 |
600–1,380 (diam.) |
|
|
Ovary length (OVL) |
248 |
187–393 |
349 |
288–394 |
283 |
243–355 |
150 |
114–182 |
300–500 (diam.) |
|
|
Ovary width (OVW) |
295 |
252–437 |
416 |
291–493 |
397 |
337–449 |
246 |
212–288 |
300–500 (diam.) |
|
|
Mehlis’ gland length (MEL) |
425 |
321–811 |
399 |
326–455 |
386 |
337–468 |
153 |
106–190 |
385a
|
|
|
Mehlis’ gland width (MEW) |
631 |
504–932 |
734 |
582–849 |
373 |
318–411 |
310 |
258–341 |
674a
|
|
|
Ant. testis length (ATL) |
645 |
470–800 |
788 |
629–932 |
564 |
430–692 |
384 |
341–455 |
1,011 (diam.)a
|
|
|
Ant. testis width (ATW) |
572 |
406–809 |
597 |
425–750 |
409 |
337–505 |
377 |
326–447 |
1,011 (diam.)a
|
|
|
Post. testis length (PTL) |
694 |
489–828 |
879 |
627–1,061 |
562 |
449–655 |
406 |
296–515 |
1,064a
|
|
|
Post. testis width (PTW) |
555 |
287–884 |
579 |
395–705 |
411 |
337–524 |
415 |
288–531 |
837a
|
|
|
Egg length (EL) |
117 |
98–132 |
114 |
108–125 |
95 |
94–96 |
113 |
104–122 |
111.3 (92–124) |
|
|
Egg width (EW) |
75 |
62–90 |
65 |
57–75 |
60 |
59–60 |
62 |
53–70 |
70.6 (65–76) |
|
|
Forebody length (FORE) |
1,937 |
1,163–3,008 |
1,369 |
1,200–1,662 |
1,408 |
1,253–1,533 |
1,218 |
985–1,600 |
1,925a
|
|
|
ODIV |
1,439 |
1,080–1,883 |
1,027 |
923–1,262 |
1,181 |
1,103–1,324 |
1,074 |
923–1,354 |
1,887a
|
|
|
OVAR |
5,876 |
4,674–6,558 |
4,461 |
3,969–5,046 |
4,856 |
4,507–5,348 |
3,043 |
2,739–3,385 |
6,740a
|
|
|
TEND |
2,742 |
1,673–4,014 |
3,653 |
3,000–4,308 |
3,081 |
2,824–3,329 |
2,059 |
1,262–2,431 |
3,370a
|
|
|
OSW/PHW |
1.21 |
1.01–1.39 |
1.29 |
1.15–1.54 |
0.97 |
0.78–1.11 |
1.41 |
1.24–1.58 |
1.27a
|
|
|
BW1/BL (%) |
9.2 |
6.32–12.3 |
12.4 |
11.2–14.3 |
11.6 |
11.2–12.5 |
16.5 |
15.0–19.8 |
15.6a
|
|
|
BW2/BL (%) |
11.5 |
7.29–16.9 |
15.4 |
13.8–17.4 |
10.3 |
9.7–10.9 |
20.7 |
18.9–24.7 |
15.6a
|
|
|
BW3/BL (%) |
17.6 |
12.0–27.7 |
17.3 |
15.2–19.5 |
13.1 |
12.4–13.6 |
23.9 |
21.5–26.9 |
20.2a
|
|
|
FORE/BL (%) |
17.5 |
9.53–31.7 |
13 |
10.9–14.8 |
14.1 |
13.4–14.9 |
19.3 |
15.5–26.3 |
14.8a
|
|
|
CW/BW3 (%) |
29.3 |
20.9–38.0 |
33.6 |
28.3–38.3 |
50.4 |
46.2–56.9 |
48.4 |
42.0–57.3 |
28.4a
|
|
|
OVID/BL (%) |
13.0 |
9.67–19.1 |
9.8 |
8.3–11.2 |
11.8 |
11.2–12.5 |
17.0 |
14.5–23.7 |
14.5a
|
|
|
OVAR/BL (%) |
52.4 |
41.0–61.0 |
42.5 |
39.1–46.2 |
48.6 |
47.7–49.7 |
48.1 |
43.5–57.0 |
29.6a
|
|
|
TEND/BL (%) |
24.1 |
17.4–35.9 |
34.6 |
31.4–37.4 |
30.9 |
30.1–31.9 |
32.3 |
22.0–35.4 |
25.9a
|
Table 3Sequence comparison of 37-collar-spined Echinostoma spp. in GenBank based on ITS region, nad1, and cox1 genes
Table 3
|
ITS region |
cox1 |
nad1 |
|
|
|
|
Between isolates (E. mekongi n. sp.) |
100 |
|
99.6 |
|
99.4–99.8 |
|
E. revolutum
|
97.6 |
E. revolutum (Southeast Asian lineage) |
90.7–91.2 |
E. revolutum (Eurasian lineage) |
85.7–86.0 |
|
|
E. robustum
|
97.9 |
E. revolutum (American lineage) |
89.4–89.9 |
E. revolutum (American lineage) |
86.3–86.6 |
|
|
E. caproni
|
97.0 |
E. miyagawai
|
90.0 |
E. miyagawai (Eurasian lineage) |
86.0–86.3 |
|
|
E. trivolvis
|
97.6 |
E. robustum
|
89.4 |
E. miyagawai (Australian lineage) |
87.4–87.7 |
|
|
E. paraensei
|
96.4 |
E. caproni
|
92.6–92.7 |
E. robustum
|
87.4–87.7 |
|
|
|
E. trivolvis
|
91.0 |
E. paraulum
|
88.0–88.2 |
|
|
|
E. paraensei
|
89.3 |
E. caproni
|
82.2–85.3 |
|
|
|
|
|
E. trivolvis
|
83.5–83.8 |
|
|
|
|
|
E. nasincovae
|
81.2–81.5 |
|
|
|
|
|
E. novaezealandense
|
86.4–86.7 |
|
|
|
|
|
E. deserticum
|
87.1–87.2 |
|
|
|
|
|
E. bolschewense
|
82.0–82.6 |
|
|
|
|
|
E. paraensei
|
85.1–85.4 |
|
|
|
|
|
E. cinetorchis
|
87.6–87.7 |
|
|
|
|
|
Echinostoma sp. IG |
79.8–80.4 |
References
- 1. Chai JY, Cho J, Chang T, Jung BK, Sohn WM. Taxonomy of Echinostoma revolutum and 37-collar-spined Echinostoma spp.: a historical review. Korean J Parasitol 2020;58:343-371.
- 2. Chai JY. Human Intestinal Flukes Chapter 2. Echinostomes. The Netherlands. Springer Nature B; 2019, pp 169-343.
- 3. Chai JY. Echinostomes in humans. In Fried B, Toledo R eds, The Biology of Echinostomes. New York, USA. Springer; 2009, pp 147-183.
- 4. Kanev I. Life-cycle, delimitation and redescription of Echinostoma revolutum (Froelich, 1802) (Trematoda: Echinostomatidae). Syst Parasitol 1994;28:125-144.
- 5. Nagataki M, Tantrawatpan C, Agatsuma T, Sugiura T, Duenngai K, Sithithaworn P, Andrews RH, Petney TN, Saijuntha W. Mitochondrial DNA sequences of 37 collar-spined echinostomes (Digenea: Echinostomatidae) in Thailand and Lao PDR reveals presence of two species: Echinostoma revolutum and E. miyagawai
. Infect Genet Evol 2015;35:56-62.
- 6. Georgieva S, Blasco-Costa I, Kostadinova A. Molecular characterization of four echinostomes (Digenea: Echinostomatidae) from birds in New Zealand, with descriptions of Echinostome novaezealandense n. sp. and Echinoparyphium poulini n. sp. Syst Parasitol 2017;94:477-497.
- 7. Kostadinova A, Herniou EA, Barrett J, Littlewood DTJ. Phylogenetic relationships of Echinostoma Rudolphi, 1809 (Digenea: Echinostomatidae) and related genera re-assessed via DNA and morphological analyses. Syst Parasitol 2003;54:159-176.
- 8. Georgieva S, Selbach C, Faltýnková A, Soldánová M, Sures B, Skirnisson K, Kostadinova A. New cryptic species of the ‘revolutum’ group of Echinostoma (Digenea: Echinostomatidae) revealed by molecular and morphological data. Parasit Vectors 2013;6:64.
- 9. Georgieva S, Faltýnková A, Brown R, Blasco-Costa I, Soldánová M, Sitko J, Scholz T, Kostadinova A.
Echinostoma ‘revolutum’ (Digenea: Echinostomatidae) species complex revisited: species delimitation based on novel molecular and morphological data gathered in Europe. Parasit Vectors 2014;7:520.
- 10. Detwiler JT, Bos DH, Minchella DJ. Revealing the secret lives of cryptic species: examining the phylogenetic relationships of echinostome parasites in North America. Mol Phylogenet Evol 2010;55:611-620.
- 11. Detwiler JT, Zajac AM, Minchella DJ, Belden LK. Revealing cryptic parasite diversity in a definitive host: echinostomes in muskrats. J Parasitol 2012;98:1148-1155.
- 12. Morgan JA, Blair D. Mitochondrial ND1 gene sequences used to identify echinostome isolates from Australia and New Zealand. Int J Parasitol 1998;28:493-502.
- 13. Saijuntha W, Sithithaworn P, Duenngai K, Kiatsopit N, Andrews RH, Petney TN. Genetic variation and relationships of four species of medically important echinostomes (Trematoda: Echinostomatidae) in South-East Asia. Infect Genet Evol 2011;11:375-381.
- 14. Noikong W, Wongsawad C, Chai JY, Saenphet S, Trudgett A. Molecular analysis of echinostome metacercariae from their second intermediate host found in a localised geographic region reveals genetic heterogeneity and possible cryptic speciation. PLoS Negl Trop Dis 2014;8:e2778.
- 15. Mohanta UK, Watanabe T, Anisuzzaman , Ohari Y, Itagaki T. Characterization of Echinostoma revolutum and Echinostoma robustum from ducks in Bangladesh based on morphology, nuclear ribosomal ITS2 and mitochondrial nad1 sequences. Parasitol Int 2019;69:1-7.
- 16. Morgan JA, Blair D. Relative merits of nuclear ribosomal internal transcribed spacers and mitochondrial CO1 and ND1 genes for distinguishing among Echinostoma species (Trematoda). Parasitology 1998;116:289-297.
- 17. Chai JY, Sohn WM, Yong TS, Eom KS, Min DY, Hoang EH, Phammasack B, Insisiengmay B, Rim HJ. Echinostome flukes recovered from humans in Khammouane Province, Lao PDR. Korean J Parasitol 2012;50:269-272.
- 18. Faltýnková A, Georgieva S, Soldánová M, Kostadinova A. A re-assessment of species diversity within the ‘revolutum’ group of Echinostoma Rudolphi, 1809 (Digenea: Echinostomatidae) in Europe. Syst Parasitol 2015;90:1-25.
- 19. Luton K, Walker D, Blair D. Comparisons of ribosomal internal transcribed spacers from two congeneric species of flukes (Platyhelminthes: Trematoda: Digenea). Mol Biochem Parasitol 1992;56:323-328.
- 20. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013;30:2725-2729.
- 21. Yamaguti S. Studies on the helminth fauna of Japan. Part 3. Avian trematodes, II. Jpn J Zool 1934;5:542-583.
- 22. Yamaguti S. Studies on the helminth fauna of Japan. Part 5. Trematodes of birds, III. Jpn J Zool 1935;6:159-182.
- 23. Sandground JH, Bonne C.
Echinostoma lindoensis n. sp., a new parasite of man in the Celebes with an account of its life history and epidemiology. Am J Trop Med Hyg 1940;20:511-536.
- 24. Kanev I, Dimitrov V, Radev V, Fried B. Redescription of Echinostoma jurini (Skvortzov, 1924) with a discussion of its identity and characteristics. Ann Naturhist Mus Wien 1995;97B:37-53.
- 25. Reddy DB, Ranganaykamma I, Venkataratnam D.
Artyfechinostomum mehrai infestation in man. J Trop Med Hyg 1964;67:58-59.
- 26. Sohn WM, Kim HJ, Yong TS, Eom KS, Jeong HG, Kim JK, Kang AR, Kim MR, Park JM, Ji SH, Sinuon M, Socheat D, Chai JY.
Echinostoma ilocanum infection in Oddar Meanchey Province, Cambodia. Korean J Parasitol 2011;49:187-190.
- 27. Sohn WM, Chai JY, Yong TS, Eom KS, Yoon CH, Sinuon M, Socheat D, Lee SH.
Echinostoma revolutum infection in children, Pursat Province, Cambodia. Emerg Infect Dis 2011;17:117-119.
- 28. Anh NTL, Phuong NT, Murrell KD, Johansen MV, Dalsgaard A, Thu LT, Chi TTK, Thamsborg SM. Animal reservoir hosts and fish-borne zoonotic trematode infections on fish farms, Vietnam. Emerg Infect Dis 2009;15:540-546.
- 29. Anh NTL, Madsen H, Dalsgaard A, Phuong NT, Thanh DTH, Murrell KD. Poultry as reservoir hosts for fishborne zoonotic trematodes in Vietnamese fish farms. Vet Parasitol 2010;169:391-394.
- 30. Chai JY, Sohn WM, Na BK, De NV.
Echinostoma revolutum: metacercariae in Filopaludina snails from Nam Dinh Province, Vietnam, and adults from experimental hamsters. Korean J Parasitol 2011;49:449-455.
- 31. Sayasone S, Tesana S, Utzinger J, Hatz C, Akkhavong K, Odermatt P. Rare human infection with the trematode Echinochasmus japonicus in Lao PDR. Parasitol Int 2009;58:106-109.
- 32. Sohn WM, Chai JY, Na BK, Yong TS, Eom KS, Park H, Min DY, Rim HJ.
Echinostoma macrorchis in Lao PDR: metacercariae in Cipangopaludina snails and adults from experimentally infected animals. Korean J Parasitol 2013;51:191-196.
- 33. Chai JY, Sohn WM, Cho J, Eom KS, Yong TS, Min DY, Hoang EH, Phammasack B, Insisiengmay B, Rim HJ.
Echinostoma ilocanum infection in two residents of Savannakhet Province, Lao PDR. Korean J Parasitol 2018;56:77-81.
- 34. Chai JY, Jung BK, Chang T, Shin H, Sohn WM, Eom KS, Yong TS, Min DY, Phommasack B, Insisiengmay B, Rim HJ. Echinostoma aegyptica (Trematoda: Echinostomatidae) infection in five riparian people in Savannakhet Province, Lao PDR. Korean J Parasitol 2020;58:67-72.
- 35. Chai JY, Chang T, Jung BK, Shin H, Sohn WM, Eom KS, Yong TS, Min DY, Phommasack B, Insisiengmay B, Rim HJ.
Echinochasmus caninus n. comb. (Trematoda: Echinostomatidae) infection in eleven riparian people in Khammouane Province, Lao PDR. Korean J Parasitol 2019;57:451-456.
- 36. Bhaibulaya M, Harinasuta C, Trirachantra S. The finding of Echinostoma revolutum infection in man in Thailand. J Med Assoc Thailand 1966;49:83.
- 37. Radomyos P, Radomyos B, Tungtrongchitr A. Multi-infection with helminths in adults from northeast Thailand as determined by post-treatment fecal examination of adult worms. Trop Med Parasitol 1994;45:133-135.
- 38. Saijuntha W, Tantrawatpan C, Sithithaworn P, Andrews RH, Petney TN. Genetic characterization of Echinostoma revolutum and Echinoparyphium recurvatum (Trematoda: Echinostomatidae) in Thailand and phylogenetic relationships with other isolates inferred by ITS1 sequence. Parasitol Res 2011;108:751-755.
- 39. Buddhachat K, Chontananarth T. Is species identification of Echinostoma revolutum using mitochondrial DNA barcoding feasible with high-resolution melting analysis? Parasitol Res 2019;118:1799-1810.
- 40. World Health Organization. Basic Laboratory Methods in Medical Parasitology. Geneva, Switzerland. World Health Organization; 1991, p 26.