Abstract
In Europe, 5 Lipoptena species have been recorded, including Lipoptena fortisetosa. This species, native to Asian countries, was described as a parasite of sika deer and its appearance in Europe dates back to more than 50 years ago. Lipoptena fortisetosa has been recently reported in Italy, sharing its hosts with Lipoptena cervi. A morpho-molecular approach was developed to determine the phylogenetic interrelationship of Italian and Asian CO1 haplotypes sequenced from Lipoptena fly individuals collected in Italy, and their DNA sequences were compared with conspecifics available in GenBank; morphological key-characters (terminalia) of L. fortisetosa were compared with the original description. Two haplotypes were recorded from Italy and assigned to L. cervi and L. fortisetosa, respectively. The latter was part of the monophyletic clade L. fortisetosa, along with 2 Central European and 2 Korean haplotypes (100% identical to one of the Korean haplotypes); moreover, Italian L. fortisetosa female terminalia were consistent with the original description of Asian individuals. Pending more in-depth investigations, this study provides a first answer to the hypothesis of the recent colonization of Italy by L. fortisetosa from Asia as we did not detect any obvious and stable morphological and molecular differences in specimens from the 2 geographical areas. The presence of the sika deer in Europe was retraced and the possible route traveled by the parasite from Asia and the eco-biological factors that may have enhanced its settlement are discussed.
-
Key words: Lipoptena fortisetosa, phylogeny, morphology, Italy, Asia
INTRODUCTION
Deer keds
Lipoptena spp. (Hippoboscidae, Lipopteninae) are blood-sucking obligate ectoparasites of almost exclusively Cervidae (deer) and Bovidae (cattle, goats, chamois, antelopes, etc.), and can occasionally bite humans [
1–
3]. Indeed, the
Lipoptena genus includes about 30 species spread worldwide [
1]: most of them occur in the Palearctic region, mainly in continental Europe and Asia, while 8 species are native to far East Asian countries. Five species have been recorded in America, with 4 of them native to this continent [
4]. Fragmentary information is available on the species accounted for African countries [
5,
6] (
Supplementary Table S1).
In severe infestations,
Lipoptena spp. may be responsible for anemia and skin lesions and may be involved in the transmission of several pathogens [
7,
8].
In Europe, 5 species of
Lipoptena have been recorded:
Lipoptena capreoli Róndani, 1878,
Lipoptena couturieri Séguy, 1935,
Lipoptena arianae Maa, 1969,
Lipoptena cervi (Linnaeus, 1758), and
Lipoptena fortisetosa Maa, 1965 [
9]; however, the presence and the geographical range of the first 3 species need to be confirmed.
Lipoptena cervi may be considered the oldest deer ked in Europe as the relationship of this species with wild ungulates dates back to more than 5,000 years ago, being found in the remains of a Late Neolithic human mummy discovered in a glacier in the Alps [
10]. It has a wide distribution in Europe and has currently been reported from more than 20 countries [
11].
Lipoptena cervi has been recorded on
Cervus elaphus,
Dama dama,
Alces alces,
Rupicapra rupicapra,
Capreolus capreolus,
Moschus moschiferus [
9] and it has the potential to transmit bacteria e.g.,
Bartonella spp.,
Borrelia spp. [
12–
14],
Anaplasma spp.,
Ehrlichia spp., and
Rickettsia spp., and protozoans, e.g.,
Babesia spp.,
Theileria spp.,
Hepatozoon spp. [
15–
17].
Lipoptena fortisetosa was originally recorded in Japan from the sika deer
Cervus nippon [
18–
20], and is considered quite restricted to this ungulate, although it has been occasionally collected from a bird
Emberiza spodocephata [
21]. It has also been reported from the Siberian roe deer
Capreolus pygargus in South Korea [
22] and in
Capreolus capreolus in Kazakhstan [
23]. This species was first reported in Europe about 50 years ago when it was found in the Czech Republic [
24] and later in the Moscow district in Russia [
25]. Afterward, from the 80s’ to date, it has been confirmed and/or recorded in 12 countries, i.e., Czech Republic, Poland, Moldavia, Germany, Switzerland, Lithuania, Romania, Austria, Belarus, Slovak Republic, Moscow-district, and Estonia [
26]. The range of
L. fortisetosa has expanded in the southern part of Europe, including Italy, where it has very recently been reported [
27,
28]. In Europe,
L. fortisetosa attacks mainly deer [
29,
30], and occasionally cattle [
31], goats, sheep [
29,
30], dogs [
32,
33], and humans, as reported in Germany [
34], Estonia [
26], and Slovakia [
35].
Lipoptena fortisetosa has been found to mechanically carry pathogens, i.e.,
Coxiella-like bacteria (CLB),
Theileria luwenshuni, and
Theileria ovis [
36]. In addition, very recently, both
L. cervi and
L. fortisetosa specimens from Poland were found positive to
Trypanosoma DNA [
37].
In Europe,
L. fortisetosa appears to share with
L. cervi approximately the same ungulate species as the host group (above listed) and roughly the same territory [
26]. However, it is unclear whether the native host (i.e., the sika deer
Cervus nippon) of
L. fortisetosa played a role in spreading the Asian species to Europe or the parasite propagated independently, as already speculated [
38], or to which extent human activities might have helped the ked expansion.
Lipoptena fortisetosa likely dispersed widely and settled in Europe quite quickly, so that possible changes in genetic constitution compared to the Asian indigenous populations may be hypothesized.
Molecular investigations coupled to morphological analysis, as well as the ecological requirements, help to provide more insight into the perspective of the integrated taxonomy concept [
39].
In order to state whether there are sharp differences between Italian and Asian individuals, we developed a morpho-molecular approach. In particular, we investigated the phylogenetic interrelationship of the mitochondrial cytochrome c oxidase subunit 1 (CO1) gene haplotypes of Lipoptena individuals sampled in Italy-analyzing them in a wider context together with the DNA sequences of conspecifics available in the NCBI GenBank and morphologically analyzed the Italian population of L. fortisetosa. We also focused on the observation of some stable key-characters (terminalia) and compared them with the original description of the species, as well as with the indigenous L. cervi species, as additional documentation. Finally, the presence of the sika deer in Europe was retraced in order to discuss the possible route traveled by the ectoparasite from Asia and the eco-biological factors that may have enhanced its settlement.
MATERIAL AND METHODS
Ethics statement, specimens and processing
All animal handling procedures followed all regional, national, and institutional guidelines.
From a total of 312
Lipoptena specimens, previously collected from 3 species of wild ruminants [
28], belonging to
Lipoptena cervi and
Lipoptena fortisetosa, the following were selected from different host species: 10 flies each from 5
Cervus elaphus hosts (total 50 specimens: 30 and 20 specimens belonging to
L. cervi and
L. fortisetosa, respectively); 10 flies from 3
Capreolus capreolus hosts (total 30 specimens, all belonging to
L. fortisetosa), 10 flies from 1
Dama dama host (total 10 specimens, all belonging to
L. fortisetosa), and frozen at −20°C, until DNA extraction. Genomic DNA was extracted individually from the abdomens using the Nucleospin Tissue kit (Macherey-Nagel, Amsterdam, Netherlands) in accordance with the manufacturer’s instructions. The extracted DNA was eluted in 50 μl of distilled water and the samples were stored at −20°C, pending molecular analysis. PCR amplifications were performed in a CFX96 thermal cycler (Bio-Rad, Hercules, California, USA.) using 10 μl of Phire Reaction Buffer 5X (Thermo Scientific, Waltham, Massachusetts, USA), 0.4 μl of dNTPs (200 μm) (Qiagen, Germantown, Maryland, USA), 1 μl of specific primer pairs (10 μm), 0.4 μl of Phire Hot Start II DNA Polymerase 1 U (Thermo Scientific), and 5 μl (approximately 100 μg) of genomic DNA per reaction. A blank control (pure water instead of genomic DNA) was included in each PCR run.
An approximately 710-bp gene fragment of
CO1 was amplified using primers LCO-1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO-2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) [
40]. The cycling parameters were: 2 min denaturing at 94°C, followed by 35 cycles of 30 sec at 94°C, 30 sec at 56°C and 60 sec at 70°C, and final extension of 7 min at 70°C. PCR products were run on 1.2% agarose gel, and positive samples purified with exonuclease I (EXO I) and thermosensitive alkaline phosphatase (FAST AP) (Fermentas, Waltham, Massachusetts, USA) enzymes, in accordance with the manufacturer’s instructions.
PCR products were directly sequenced in both directions using the ABI PRIMS BygDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, California, USA) with the same primers as the respective PCR reactions, in accordance with the manufacturer’s instructions. The sequences obtained were determined using an ABI PRISM 3130 Genetic Analyser (Applied Biosystems), chromatograms were inspected by eye using FinchTV (
https://digitalworldbiology.com/FinchTV) and primer regions plus bad-quality regions were removed. Once the sequences were cleaned up, each sequence was compared with the
Lipoptena spp. homologous nucleotide sequences available in the GenBank database using the BLAST program (Basic Local Alignment Search Tool;
https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_L+OC=blasthome). The 19 sequences having the highest percent similarity with our sequences and labelled as
Lipoptena CO1 in Genbank were then sampled and gathered in a FASTA file with our own sequences. The new sequence dataset was aligned using the CLUSTALW implementation of BIOEDIT, version 7.0.5 (
http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and the alignment adjusted manually, if necessary. Once the sequences were aligned, the absence of stop codons was checked. Phylogenetic analysis of the obtained sequences and homologous sequences from GenBank were performed using the maximum likelihood method in MEGA, version 7.0.9 (
https://www.megasoftware.net). Bootstrap confidence values for the branching reliability were calculated with 10,000 replicates.
All specimens of
L. fortisetosa and
L. cervi intended for molecular analysis were morphologically identified, based on Maa’s original description [
19] and a recent taxonomic key [
9]. Other specimens were processed for Scanning Electron Microscope (SEM) observations, according to the procedures previously described [
28] to further examine male and female terminalia. Description of terminalia features follows the terminology and nomenclature reported by Maa and Peterson [
41].
RESULTS
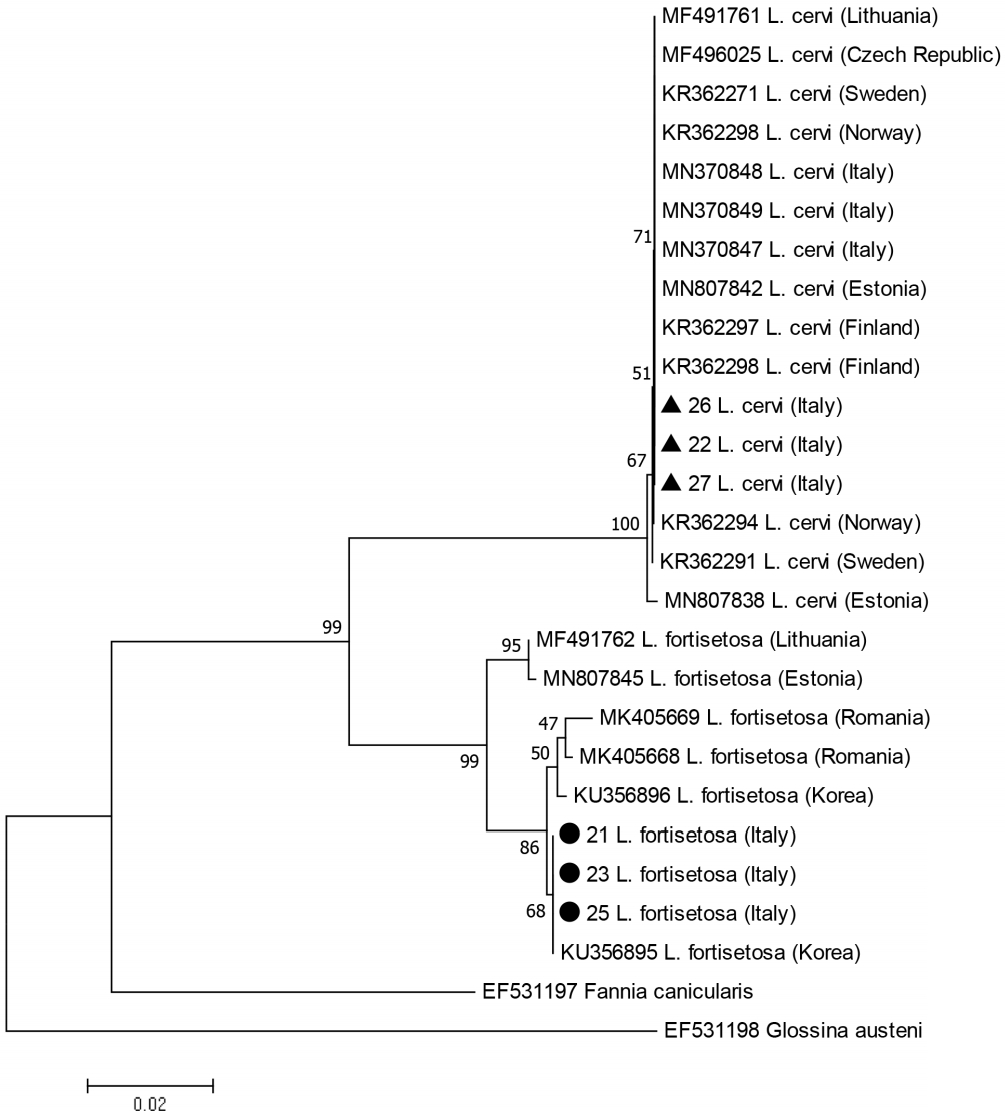
A total 60/90 (66.7%) of the specimens provided good quality PCR fragments and sequences for the CO1 gene. After alignment with the homologous sequences of Lipoptena spp. available in GenBank, 2 sets of sequences were identified, one with the mean percentage of identity of 89% with L. cervi and 92% with L. fortisetosa, and one with 97% with L. cervi and 86% with L. fortisetosa. Two haplotypes were recorded from the 60 specimens sequenced in the present study and the phylogenetic analysis confirmed that these 2 haplotypes belonged to L. cervi and L. fortisetosa, respectively. The data matrix comprised 14 haplotypes of L. cervi (one haplotype from the present study, 13 downloaded from GenBank) and 7 haplotypes of L. fortisetosa (one haplotype from the present study, 6 downloaded). The genetic distances ranged from 0.081–0.084 for L. cervi group sequences and 0.003–0.018 for L. fortisetosa group sequences.
While the clade
L. fortisetosa appears to be monophyletic, the internal 2-subclade structure of this clade is poorly supported (low bootstrap values) as expected within the species. We will thus note essentially the following points: the single haplotype recorded from the Italian
L. fortisetosa specimens was closely related to 2 Central European haplotypes and 100% identical to one of the 2 haplotypes (KU356895) found in Korea (
Fig. 1).
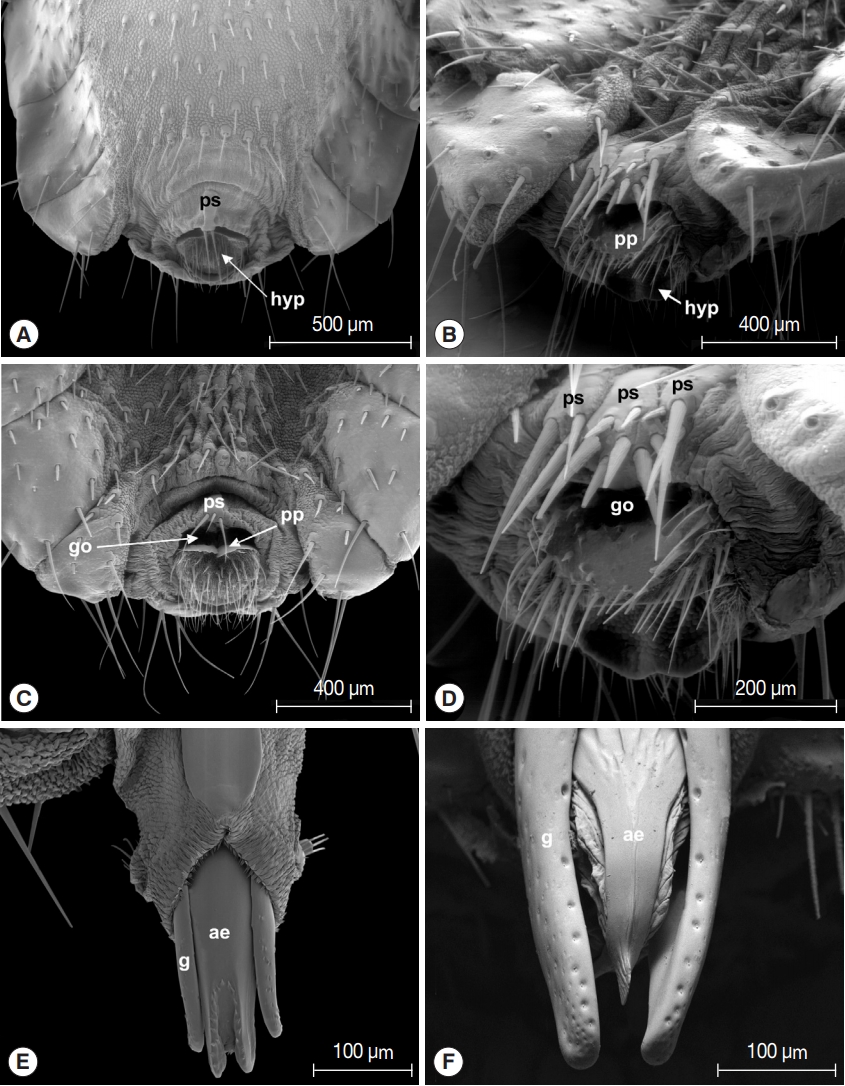
Morphological investigations showed that female terminalia of
L. fortisetosa are characterized by a typical pregenital sclerite that is peg-like and bears 2 or 3 strong bristles (
Fig. 2A, C). The pregenital plate is elongated and lozenge-shaped, and the underlying hypoproct is covered by several bristles interspersed with an area densely hairy. The genital opening is clearly visible between the pregenital sclerite and the pregenital plate (
Fig. 2C). Female terminalia of
L. cervi showed the presence of 3 pregenital sclerites bearing several differently sized bristles (
Fig. 2B, D). The central sclerite is bigger and with more numerous setae than the external 2; the pregenital plate shows many series of long setae arranged in the distal portion, while the hypoproct is completely bare. Male terminalia of
L. fortisetosa (
Fig. 2E) consist of 2 well-sclerotized and slender gonopods that guide the aedeagus. This latter is wider in the proximal part and ends with a bilobate tip provided with spines. In
L. cervi, the gonopods (
Fig. 2F) are similar to those of
L. fortisetosa. However, the aedeagus is membranous in the proximal and lateral parts, while in the middle it is formed by 2 fused and sclerotized strips ending in a ridge tip. Morphological observations of
L. cervi and
L. fortisetosa strengthened the strong diversity between these 2 species [
28] but, more importantly, demonstrate that the features of
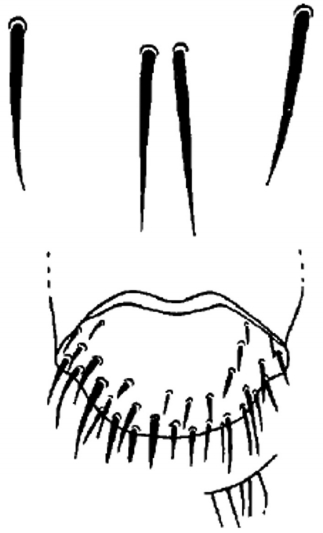
L. fortisetosa female terminalia are consistent with the original description by Maa [
18] (
Fig. 3).
Taking into account that terminalia are considered stable features that allow a correct morphological identification at species-specific level in Diptera [
42], including hippoboscids [
19,
43], this supports the above confirmation that the boundaries between the 2 species (
L. fortisetosa and
L. cervi) are correct based on morphology and molecular data. Therefore, we can conclude that the individuals of which we only have the sequences (GenBank) belong to the same species.
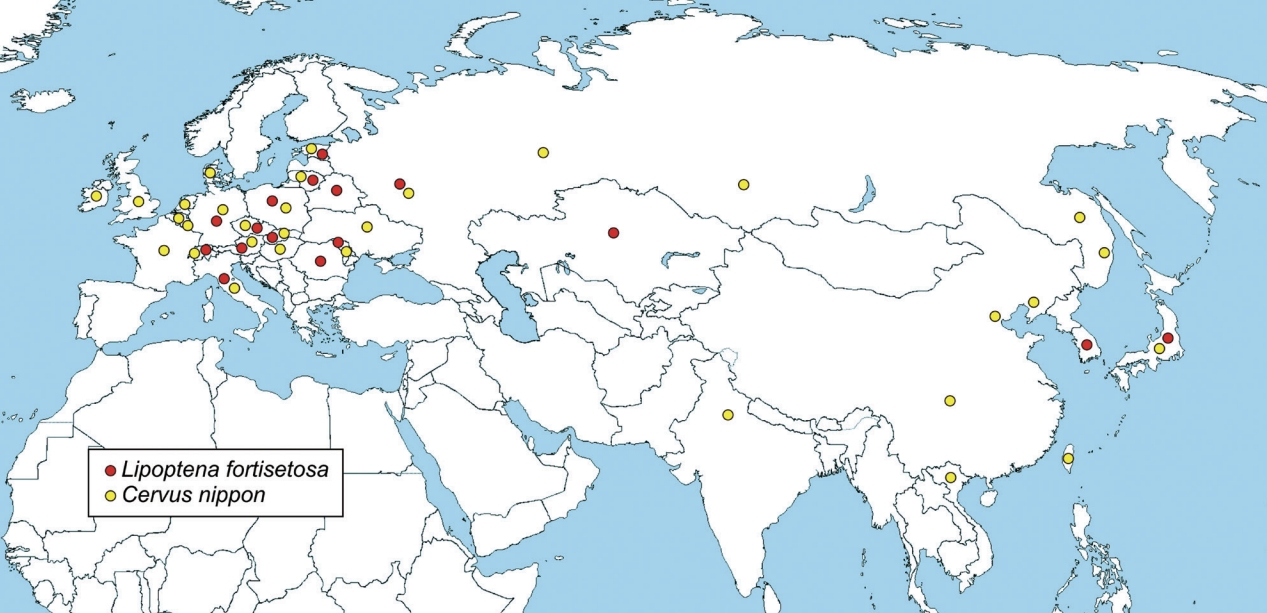
Lipoptena fortisetosa has been introduced in Europe probably with
Cervus nippon, the sika deer [
28,
33,
44], during the last 150 years of restocking of deer in the Continent, apparently, since 1893, and probably a number of times [
44]. Sika deer has successfully settled in the European fauna thanks to its high potential to compete with autochthonous species and readiness to hybridize with native red deer, as demonstrated by the presence of hybrids of sika with red deer in several countries [
45–
49]. Sika deer (or hybrids) is currently present in 20 European countries, including Italy, where it was recently reported [
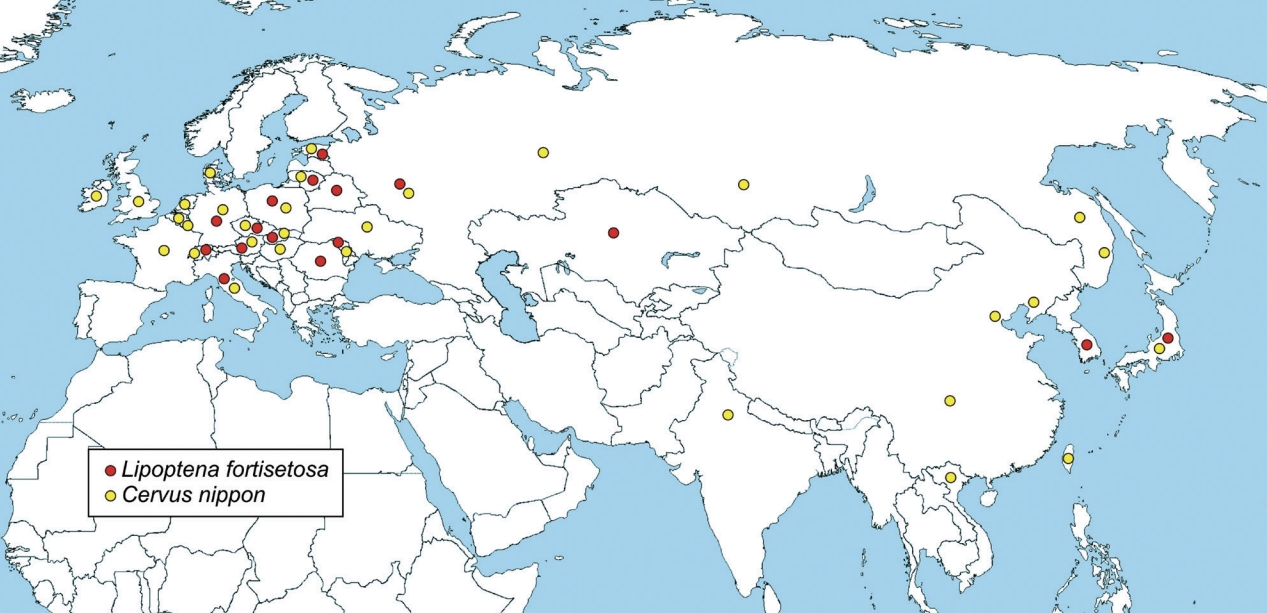
50]. Spreading through Europe, sika deer has likely carried and disseminated its ectoparasites, including
L. fortisetosa that is currently recorded in 13 European countries (
Fig. 4), Italy included [
27].
DISCUSSION
According to the present results, while
L. cervi confirms previous findings [
9], we cannot exclude a scenario depicting a recent colonization(s) of
L. fortisetosa in Europe. According to the latest studies, the hypothesis of geographically distinct
CO1 lineages should be rejected at the Central Europe level [
26]. Our study suggests that this hypothesis should also be rejected at a global level. However, we were unable to determine the time of colonization or to identify the actual host that introduced
L. fortisetosa from Asia to Europe. In fact, this parasite has been collected from many different cervids, e.g., red deer (
Cervus elaphus) [
27,
28], Manchurian elk (
Cervus elaphus xanthopygus), Maral red deer (
Cervus elaphus maral), fallow deer (
Dama dama) [
34], Korean water deer (
Hydropotes inermis) [
36], roe deer (
Capreolus capreolus) [
23], Siberian roe deer (
Capreolus pygargus) [
22], both in Europe and in Asia. The possibility of recent colonization by
L. fortisetosa specimens from cervids purchased for the restocking of deer farms, or in captive hosts in the Oriental region transferred to fenced areas for recreational or conservation purposes, cannot be excluded. We also suggest, as a minor hypothesis, the relatively recent migration to Italy of sika or hybrid individuals from neighboring countries that may have transmitted the ectoparasite joining red deer groups.
This study doesn’t reject the hypothesis of the recent colonization of Italy by
L. fortisetosa from Asia as no obvious and stable morphological and molecular differences were observed in the populations from the 2 regions. However, such a hypothesis requires further study, including a straightforward analysis of numerous specimens from Asian and European countries. In line with other authors’ suggestions [
26], population genetic analyses of Asian and European keds’ populations are required to evaluate whether the colonization scenario is likely or not. Moreover, wider investigations aimed at comparing genetic make-up of populations from different countries, host and parasite distribution, together with morphological differentiation, might resolve phylogenetic relationships of this neglected group within a desirable integrated taxonomic framework as applied in other groups of insects [
51,
52].
Due to the ability of
L. fortisetosa to parasitize a wide range of homeothermic animals, there are no obvious limitations to its further expansion. Given such possibility, the potential for transmitting pathogens, and the frequent attacks reported among humans [
26], more in-depth investigations are required on
Lipoptena species with a One Health perspective.
Notes
-
CONFLICT OF INTEREST
The authors don’t have any conflicts of interest to declare.
Supplementary Information
Supplementary Table S1.
Species of the genus Lipoptena spp. described in the world with their distribution and hosts [
1,
4-
6,
19,
43]
ACKNOWLEDGMENT
This research was partially funded by the “Fondazione Cassa di Risparmio di Firenze, Italy”.
Fig. 1Phylogenetic topology based on the analysis of the maximum likehood of the partial CO1 gene sequences from Lipoptena individuals from the present study and Lipoptena sequences available from GenBank. Labels include accession numbers, species identity and country origin. The 2 haplotypes found from the present study are labelled with a black triangle for Lipoptena cervi and a black circle for Lipoptena fortisetosa. Fannia cunicularis and Glossina austeni sequences were used as outgroups. The percentage of trees in which the associated individuals clustered together is shown next to the branches.

Fig. 2Terminalia of Lipoptena fortisetosa (A&C, female; E, male) and Lipoptena cervi (B&D, female; F, male). ps, pregenital sclerite; hyp, hypoproct; go, genital opening; pp, pregenital plate; ae, aedeagus; g, gonopod.

Fig. 3Female terminalia of
Lipoptena fortisetosa, drawing from Maa [
18].

Fig. 4Distribution map of
Lipoptena fortisetosa (red dot) and
Cervus nippon (sika deer) (yellow dot). Data from different sources [
11,
19,
23,
26,
44,
49,
50].
References
- 1. Dick CW. Checklist of world Hippoboscidae (Diptera: Hippoboscoidea). Chicago, USA. Department of Zoology, Field Museum of Natural History; 2006, pp 1-7.
- 2. Kortet R, Härkönen L, Hokkanen P, Härkönen S, Kaitala A, Kaunisto S, Laaksonen S, Kekäläinen J, Ylönen H. Experiments on the ectoparasitic deer ked that often attacks humans; preferences for body parts, colour and temperature. Bull Entomol Res 2010;100:279-285. https://doi.org/10.1017/S0007485309990277
- 3. Soós Á, Hůrka K. Family hippoboscidae. In Soós Á, Papp L eds, Catalogue of Palaearctic Diptera. 11:Budapest, Hungary. Akademiai Kaido; 1986, pp 215-226.
- 4. Skvarla MJ, Machtinger ET. Deer keds (Diptera: Hippoboscidae: Lipoptena and Neolipoptena) in the United States and Canada: new state and county records, pathogen records, and an illustrated key to species. J Med Entomol 2019;56:744-760. https://doi.org/10.1093/jme/tjy238
- 5. GBIF.org. GBIF Home Page [Internet]; [cited 2020 September 20]. Available from: https://www.gbif.org
- 6. Visagie EJ. A new species of Lipoptena (Diptera: Hippoboscidae) from Southern Africa. Onderstepoort J Vet Res 1992;59:293-302.
- 7. Härkönen S, Laine M, Vornanen M, Reunala T. Deer ked (Lipoptena cervi) dermatitis in humans−an increasing nuisance in Finland. Alces 2009;45:73-79.
- 8. Lazăr M, Iacob OC, Solcan C, Paşca SA, Lazăr R, Boişteanu PC. The first report of massive infestation with Lipoptena cervi (Diptera: Hippoboscidae) in roe deer (Capreolus capreolus) in Iasi county, N-E of Romania. Arq Bras Med Vet Zootec 2017;69:293-298. https://doi.org/10.1590/1678-4162-8612
- 9. Salvetti M, Bianchi A, Marangi M, Barlaam M, Giacomelli S, Bertoletti I, Roy L, Giangaspero A. Deer keds on wild ungulates in northern Italy, with a taxonomic key for the identification of Lipoptena spp. of Europe. Med Vet Entomol 2020;34:74-85. https://doi.org/10.1111/mve.12411
- 10. Gothe R, Schöl H. Deer keds (Lipoptena cervi) in the accompanying equipment of the late Neolithic human mummy from the Similaun, South Tyrol. Parasitol Res 1994;80:81-83. https://doi.org/10.1007/BF00932630
- 11. Fauna Europaea. All European Animal Species Online [Internet]; [cited 2020 September 20]. Available from: https://fauna-eu.org
- 12. Dehio C, Sauder U, Hiestand R. Isolation of Bartonella schoenbuchensis from Lipoptena cervi, a blood-sucking arthropod causing deer ked dermatitis. J Clin Microbiol 2004;42:5320-5323. https://doi.org/10.1128/JCM.42.11.5320-5323.2004
- 13. Duodu S, Madslien K, Hjelm E, Molin Y, Paziewska-Harris A, Harris PD, Colquhoun DJ, Ytrehus B. Bartonella infections in deer Keds (Lipoptena cervi) and moose (Alces alces) in Norway. Appl Environ Microbiol 2013;79:322-327. https://doi.org/10.1128/AEM.02632-12
- 14. Szewczyk T, Werszko J, Steiner-Bogdaszewska Ż, Jeżewski W, Laskowski Z, Karbowiak G. Molecular detection of Bartonella spp. in deer ked (Lipoptena cervi) in Poland. Parasit Vectors 2017;10:487. https://doi.org/10.1186/s13071-017-2413-0
- 15. Chomel BB, Boulouis HJ, Breitschwerdt EB, Kasten RW, Vayssier-Taussat M, Birtles RJ, Koehler JE, Dehio C. Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet Res 2009;40:29. https://doi.org/10.1051/vetres/2009011
- 16. Hornok S, de la Fuente J, Biró N, Fernández de Mera IG, Meli ML, Elek V, Gönczi E, Meili T, Tánczos B, Farkas R, Lutz H, Hofmann-Lehmann R. First molecular evidence of Anaplasma ovis and Rickettsia spp. in keds (Diptera: Hippoboscidae) of sheep and wild ruminants. Vector Borne Zoonotic Dis 2011;11:1319-1321. https://doi.org/10.1089/vbz.2011.0649
- 17. Kelsey A, Finch J. Deer ked: a Lyme-carrying ectoparasite on the move. Cutis 2018;102:121-122.
- 18. Maa TC. A synopsis of the Lipopteninae (Diptera: Hippoboscidae). J Med Entomol 1965;2:233-248. https://doi.org/10.1093/jmedent/2.3.233
- 19. Maa TC. A revised checklist and concise host index of Hippoboscidae (Diptera). Pacific Insects Monograph 1969;20:261-269.
- 20. Mogi M. A new species of Lipoptena (Diptera, Hippoboscidae) from the Japanese deer. Kontyû 1975;43:387-392.
- 21. Yamauchi T, Tsurumi M, Kataoka N. Distributional records of Lipoptena species (Diptera: Hippoboscidae) in Japan and Jeju-do, Korea. Med Entomol Zoo 2009;60:131-133. https://doi.org/10.7601/mez.60.131
- 22. Choi CY, Lee S, Moon KH, Kang CW, Yun YM. New Record of Lipoptena fortisetosa (Diptera: Hippoboscidae) collected from Siberian roe deer on Jeju Island, Korea. J Med Entomol 2013;50:1173-1177. https://doi.org/10.1603/me12150
- 23. Edwards SJ, Hood MW, Shaw JH, Rayburn JD, Kirby MD, Hanfman DT, Zidar JA. Index-Catalogue of Medical and Veterinary Zoology. Supplement 21, Part 5: Parasite-Subject Catalogue. Parasites: Arthropoda and Miscellaneous Phyla. Washington DC, USA. USDA Government Printing Office; 1978, p 246.
- 24. Theodor O. Lipoptena parvula, n. sp., eine neue Art aus der Tschechoslowakei (Diptera, Hippoboscidae). Acta Entomol Mus Natl Pragae 1965;37:275-278.
- 25. Grunin KJ. Hippoboscidae-krovososki. In Bej-Bienko GJ ed, Opredlitl nasekomych evropejskoj czasti SSSR T. 5:Nauka. Leningrad; 1970, pp 979-987 (in Russian).
- 26. Kurina O, Kirik H, Õunap H, Õunap E. The northernmost record of a blood-sucking ectoparasite, Lipoptena fortisetosa Maa (Diptera: Hippoboscidae), in Estonia. Biodivers Data J 2019;7:e47857. https://doi.org/10.3897/BDJ.7.e47857
- 27. Andreani A, Belcari A, Sacchetti P, Ponzetta MP. Occurrence of a new parasite of the ungulates in Italy: Lipoptena fortisetosa (Diptera: Hippoboscidae). Atti del IV Congresso Nazionale di Ecopatologia della Fauna. 2017 October 11–13; Società Italiana: SIEF di Ecopatologia della Fauna; 2017. 36.
- 28. Andreani A, Sacchetti P, Belcari A. Comparative morphology of the deer ked Lipoptena fortisetosa first recorded from Italy. Med Vet Entomol 2019;33:140-153. https://doi.org/10.1111/mve.12342
- 29. Büttiker W. Die Lausfliegen der Schweitz (Diptera, Hippoboscidae): mit Bestimmungsschlüssel. Neuchâtel, Switzerland. Centre Suisse de cartographie de la faune; 1994, pp 1-117 (in German).
- 30. Schedl W. Beitrag zur Lausfliegen-Fauna an Säugetieren von Kärnten und anderen Bundesländern Österreichs (Insecta: Diptera, Hippoboscidae). Linzer biol Beitr 2018;50:1283-1293. (in Deutsch).
- 31. Metelitsa AK, Veselkin GA. Parasitism of the louse fly Lipoptena fortisetosa on cattle. Parazitologiia 1989;23:276-277. [In Russian].
- 32. Sokół R, Gałęcki R. Prevalence of keds on city dogs in Central Poland. Med Vet Entomol 2017;31:114-116. https://doi.org/10.1111/mve.12209
- 33. Mihalca AD, Păstrav IR, Sándor AD, Deak G, Gherman CM, Sarmaşi A, Votýpka J. First report of the dog louse fly Hippobosca longipennis in Romania. Med Vet Entomol 2019;33:530-535.
- 34. Schumann H, Messner B. Erstnachweis von Lipoptena fortisetosa Maa, 1965 in Deutschland (Dipt., Hippoboscidae). Entomol Nachr Ber 1993;37:247-249.
- 35. Oboňa J, Sychra O, Greš S, Heřman P, Manko P, Roháček J, Šestáková A, Šlapák J, Hromada M. A revised annotated checklist of louse flies (Diptera, Hippoboscidae) from Slovakia. ZooKeys 2019;862:129-152. https://doi.org/10.3897/zookeys.862.25992
- 36. Lee SH, Kim KT, Kwon OD, Ock Y, Kim T, Choi D, Kwak D. Novel detection of Coxiella spp., Theileria luwenshuni, and T. ovis endosymbionts in deer keds (Lipoptena fortisetosa). PLoS One 2016;11:e0156727. https://doi.org/10.1371/journal.pone.0156727
- 37. Werszko J, Steiner-Bogdaszewska Ż, Jeżewski W, Szewczyk T, Kuryło G, Wołkowycki M, Wróblewski P, Karbowiak G. Molecular detection of Trypanosoma spp. in Lipoptena cervi and Lipoptena fortisetosa (Diptera: Hippoboscidae) and their potential role in the transmission of pathogens. Parasitology 2020;1-26. https://doi.org/10.1017/S0031182020001584
- 38. Kowal J, Nosal P, Kornaś S, Wajdzik M, Matysek M, Basiaga M. Biodiversity and importance of hippoboscids infection in cervids. Med Weter 2016;72:745-749.
- 39. Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH. Integrative taxonomy: a multisource approach to exploring biodiversity. Annu Rev Entomol 2010;55:421-438. https://doi.org/10.1146/annurev-ento-112408-085432
- 40. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome CO1idase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 1994;3:294-299.
- 41. Maa TC, Peterson BV. Hippoboscidae. In McAlpine JF, Peterson BV, Shewell GE, Teskey HJ, Vockeroth JR, Wood DM eds, Manual of Nearctic Diptera, Vol. II Mograph 28. Ottawa, ON, Canada. Research Branch, Agriculture Canada; 1987, pp 1271-1281.
- 42. McAlpine JF. Morphology and terminology – Adults. In McAlpine JF, Peterson BV, Shewell GE, Teskey HJ, Vockeroth JR, Wood DM eds, Manual of Nearctic Diptera, Vol. I. Mograph 27. Ottawa, ON, Canada. Research Branch, Agriculture Canada; 1981, pp 9-63.
- 43. Bequaert J. A monograph of the Melophaginae, or ked-flies, of sheep, goats, deer and antelopes (Diptera, Hippoboscidae). Entomol Americana 1942;22:1-220.
- 44. Bartoš L. Sika deer in continental Europe. In McCullough DR, Takatsuki S, Kaji K eds, Sika Deer: Biology and Management of Native and Introduced Populations. Tokyo, Japan. Springer; 2009, pp 573-594.
- 45. Lowe VPW, Gardiner AS. Hybridization between Red deer (Cervus elaphus) and Sika deer (Cervus nippon) with particular reference to stocks in N.W. England. J Zool 1975;177:553-566. https://doi.org/10.1111/j.1469-7998.1975.tb02259.x
- 46. Goodman SJ, Barton NH, Swanson G, Abernethy K, Pemberton JM. Introgression through rare hybridization: a genetic study of a hybrid zone between Red and Sika deer (Genus Cervus) in Argyll, Scotland. Genetics 1999;152:355-371.
- 47. Carden RF, Carlin CM, Marnell F, Mcelholm D, Hetherington J, Gammell MP. Distribution and range expansion of deer in Ireland. Mammal Rev 2011;41:313-325. https://doi.org/10.1111/j.1365-2907.2010.00170.x
- 48. Biedrzycka A, Solarz W, Okarma H. Hybridization between native and introduced species of deer in Eastern Europe. J Mammal 2012;93:1331-1341. https://doi.org/10.1644/11-MAMM-A-022.1
- 49. Ferri M, Fontana R, Lanzi A, Armaroli E, Peloso F, Musarò C, Andina L, Allegri M, Adorni PL, Gelmini L, Barančekovà M, Levrini M, De Pietri A, Berti E. Some Sika deer (Cervus nippon) recently hunted and spotted free-ranging in the Emilia-Romagna’s region (and out of it) question the management of Italian Red deer (Cervus elaphus) population. X Congresso Italiano di Teriologia, Hystrix 2016;27(suppl):100.
- 50. Raganella Pelliccioni E, Riga F, Toso S. Linee Guida per la gestione degli Ungulati: Cervidi e Bovidi. Roma, Italy. ISPRA Press; 2013, pp 1-220 (in Italiano).
- 51. Kim H, Hoelmer KA, Lee W, Kwon YD, Lee S. Molecular and morphological identification of the soybean aphid and other aphis species on the primary host Rhamnus davurica in Asia. Ann Entomol Soc Am 2010;103:532-543. https://doi.org/10.1603/AN09166
- 52. Hendrichs J, Vera T, De Meyer M, Clarke AR. Resolving cryptic species complexes of major tephritid pests. Zookeys 2015;540:5-39. https://doi.org/10.3897/zookeys.540.9656
 , Marianna Marangi2, Alessandra Barlaam2, Maria Paola Ponzetta1, Lise Roy3,*
, Marianna Marangi2, Alessandra Barlaam2, Maria Paola Ponzetta1, Lise Roy3,* , Antonio Belcari1
, Antonio Belcari1 , Patrizia Sacchetti1
, Patrizia Sacchetti1