Abstract
Concerns about foodborne illnesses caused by Kudoa septempunctata are steadily growing, but reports of K. septempunctata in clinical and food specimens related to food poisoning in Korea are limited. This study aimed to genetically identify K. septempunctata in patients with acute diarrhea and in clinical and food samples related to food poisoning caused by sashimi consumption. Both real-time and nested polymerase chain reaction assays were performed to detect K. septempunctata 18S and 28S rDNA genes in the stools of 348 patients with acute diarrhea, 11 samples (6 stool and 5 rectal swab samples) from patients with food poisoning, and 2 raw Paralichthys olivaceus samples collected from a restaurant where a food poisoning incident occurred. K. septempunctata was identified in 5 clinical specimens (4 stools and 1 rectal swab) and 1 P. olivaceus sashimi sample. All detected K. septempunctata were of genotype ST3. This is the first study to identify K. septempunctata in both patients and food samples with epidemiological relevance in Korea, providing evidence that it is a pathogen that causes food poisoning. Also, this is the first study to confirm the presence of K. septempunctata genes in rectal swabs. Despite continuing suspected occurrences of Kudoa foodborne outbreaks, the rate of identification of K. septempunctata is very low. One reason for this is the limitation in obtaining stool and vomit samples for the diagnosis of Kudoa infection. We strongly suggest the inclusion of rectal swabs among the diagnostic specimens for Kudoa food poisoning.
-
Key words: Kudoa septempunctata, Paralichthys olivaceus, food poisoning, olive flounder, ST3
Introduction
The genus
Kudoa is a myxosporean parasite that infects the somatic muscles, brain, pericardium, and digestive tract of various marine fishes [
1,
2]. Certain species of
Kudoa cause postmortem liquefaction known as “ jelly meat”, which reduces the commercial value of fish [
3,
4]. A study on
Kudoa septempunctata as a causative organism of food poisoning was performed in Japan [
5]. In that study, 130 of the 158 cases of unknown foodborne illness reported in 2010 were associated with the consumption of raw
Paralichthys olivaceus (olive flounder), and
K. septempunctata genes and spores were identified in the
P. olivaceus remnants of patient meals. The Ministry of Health, Labor and Welfare of Japan announced
K. septempunctata as a potential cause of food poisoning, and tests on domestic and imported
P. olivaceus are further being strengthened [
6]. Similarly, after the reports of the presence of
K. septempunctata in cultured
P. olivaceus in Jeju, Korea [
4,
7,
8], the Ministry of Food and Drug Safety of Korea proposed and circulated a method for detecting
K. septempunctata in
P. olivaceus in 2013. Marine aquaculture has rapidly become an essential component of the global marine food supply, accounting for 44% of the global fish production in 2014. That year, Korea ranked 7th among the world’s top 25 aquaculture producers [
9]. In Korea,
P. olivaceus accounted for 46.2% (37,240 metric tons) of the total fish farming yield (80,530 metric tons) in 2018 [
10]. Foodborne outbreaks associated with
P. olivaceus infected with
K. septempunctata are crucial because they can lead to economic losses.
K. septempunctata has 3 different genotypes: ST1, ST2, and ST3. The genotypes are determined by a combination of 2 mitochondrial genes, namely, cytochrome
c oxidase subunit 1 (
cox1) and large subunit rRNA (
rnl). There are 3 alleles (
cox1-1,
cox1-2, and
cox1-3) for
cox1, which differ at 7 single nucleotide polymorphisms (SNPs). Whereas there are 2 alleles (
rnl-1 and
rnl-2) for
rnl, which differ at 2 SNPs [
11]. Studies have shown that
K. septempunctata found in Japan mainly has the ST1 or ST2 genotype, whereas ST3 genotype is found in Korea [
11–
14].
Korean and Japanese food cultures involve frequent consumption of raw fish in the forms of sashimi and sushi. Since these cuisines are spreading around the world, identification of food poisoning caused by
K. septempunctata is important from the public health point of view. In 2015, the Korea Disease Control and Prevention Agency (KDCA) announced foodborne illness outbreaks caused by
K. septempunctata [
15] but classified them as “other infections” that cause food poisoning [
16]. This is due to the continuing controversy about whether
K. septempunctata causes food poisoning, resulting from conflicting results on its pathogenicity in experiments using mice [
5,
12,
13] and the possibility of confusion with toxin-type food poisoning (
Staphylococcus aureus or
Bacillus cereus) [
17]. It is therefore important to detect
K. septempunctata in feces or vomit samples obtained from patients with food poisoning. However, to date, only 2 studies have been performed in Korea [
18,
19]. In addition, although the KDCA has been monitoring 20 food poisoning pathogens for patients with acute diarrhea at cooperative hospitals nationwide through the Enteric Pathogens Active Surveillance Network (Enter-Net) every year,
K. septempunctata has not been included in this survey. Thus, information available on the status of
K. septempunctata infection in Korea is limited.
The present study investigated the infection status of K. septempunctata by detecting specific genes (18S and 28S rDNA) in stool samples from patients admitted with acute diarrhea to hospitals participating in the Enter-Net system in Busan. Patients in whom food poisoning was caused due to the consumption of seafood were included. Furthermore, raw P. olivaceus samples were collected from restaurants where food poisoning incidents had occurred. These samples were also used for the genetic identification and genotyping of K. septempunctata to investigate its potential as a food poisoning pathogen and emphasize the importance of managing and preventing such outbreaks.
Materials and Methods
Collection of samples
From November 2019 to October 2020, 348 stool samples of patients with acute diarrhea were collected from 5 hospitals in Busan. In addition, 11 samples (6 stool and 5 rectal swab samples) were collected from 6 food poisoning outbreaks that had occurred due to the consumption of seafood. Two samples of P. olivaceus sashimi were collected from a restaurant where a food poisoning incident had occurred. Samples were collected at public health centers and the Environmental Sanitation Department of district offices.
Sample preprocessing and DNA extraction
The collected stool, rectal swabs (Transystem 132C, Copan, Brescia, Italy), and P. olivaceus sashimi samples were stored in a cool box maintained at 4–8°C and transported to our laboratory within 4 h of collection. The samples were stored at 4°C, and nucleic acid amplification assays were performed as early as possible within 48 h of the arrival of the samples.
For DNA extraction from the rectal swabs, the sample was cut with sterile scissors, placed in a 15 ml conical tube, and vortexed with sterile glass beads and 500 μl of 0.1 M phosphate buffered saline (PBS, pH 7.4, Sigma-Aldrich, St. Louis, MO, USA) for 10 min.
P. olivaceus sashimi sample extraction was performed as per the guidelines mentioned in the Manual for Detection of Foodborne Pathogens at Outbreaks [
20]. In brief, (A) the surface of the sashimi was scratched in at least 5 places to obtain 1 g sample; (B) the collected sample was placed on a mesh of approximately 200 μm, to which 3 ml of PBS was added, (C) light mashing was performed; (D) the resultant solution collected through the mesh was centrifuged at 1,500 g for 15 min at 4°C, and (E) the solute was discarded and 1 ml of PBS was added to the precipitate.
DNA was extracted from 250 μl of the prepared homogenized solution and 0.25 g of stool. DNA extraction was performed using the QIAcube and QIAamp PowerFecal DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Genetic identification and genotype analysis
For screening of
K. septempunctata infection, all DNA extracts were subjected to real-time polymerase chain reaction (PCR) assay on the ABI 7500 Fast System (Applied Biosystems, Foster City, CA, USA) using the PowerChek
Kudoa Real-time PCR kit (KogeneBiotech, Seoul, Korea). After obtaining specific fluorescence amplification curves, nested PCR was performed to detect the 18S and 28S ribosomal DNAs. Primers of 2
K. septempunctata mitochondrial genes (
cox1 and
rnl) were used for genotype analysis. The primer sets and references are presented in
Table 1. PCR was performed using the ProFlex PCR System (Applied Biosystems) with AccuPower PCR Premix (Bioneer, Daejeon, Korea) according to the manufacturer’s instructions.
The amplified products were visualized on the QIAxcel Advanced System (Qiagen) and sequenced bidirectionally by Bionics Co. (Bionics Co., Seoul, Korea). The sequences were aligned by applying the BioEdit sequence alignment program (
https://bioedit.software.informer.com/7.2) and analyzed using NCBI-BLAST (
https://www.ncbi.nlm.nih.gov/Blast). For genotype analysis, sequences were compared with the standard strain using Clustal W in BioEdit (
Table 2).
Statistical analyses were performed using SPSS version 22.0 (IBM, Armonk, NY, USA). Mean and standard deviation were calculated for the quantitative variables, and percentages were calculated for the qualitative variables. Independent sample t-test and chi-square test were applied to determine the correlation between the K. septempunctata 18S and 28S rDNA genes and each variable. Statistical significance was set at P<0.05.
Results
Prevalence and genetic identification of K. septempunctata
Real-time PCR for the screening of K. septempunctata was performed on the stool samples of 348 patients with acute diarrhea, 11 samples (6 stool and 5 rectal swab samples) from patients with food poisoning, and 2 raw P. olivaceus samples collected from a restaurant where a food poisoning incident had occurred. Real-time PCR amplification curves were observed in 4 patients with acute diarrhea, 5 patients with food poisoning, and 2 raw P. olivaceus samples, for which 18S and 28S rDNA nested PCR were performed. PCR products were sequenced and compared with the K. septempunctata gene for 18S rDNA (accession no. AB731754.1) and 28S rDNA (accession no. AB731755.1). Finally, 5 patients with food poisoning and 1 raw P. olivaceus sample were determined to be positive for K. septempunctata.
Epidemiological characteristics of 11 patients in whom food poisoning occurred due to the consumption of seafood in Busan during the same period are summarized in
Table 3. Of the 11 patients examined, the average incubation period per patient was 4.7 h, with 81.8% of the patients experiencing diarrhea and abdominal pain and 54.5% experiencing vomiting. The detection rate of
K. septempunctata was 45.5% (5 patients), with all patients reported to have consumed
P. olivaceus sashimi. Three out of these 5 (60%) patients experienced multiple symptoms concurrently, such as diarrhea, vomiting, and abdominal pain. One out of the 5 patients experienced diarrhea and vomiting, and the other one experienced only abdominal pain. The time interval between food intake and fecal sample collection ranged from 16 to 58 h;
K. septempunctata was not detected in the rectal swabs collected 58 h later. Of the fecal samples collected from 11 patients, 6 were stool samples (54.5%). The detection rate of
K. septempunctata via PCR in stool samples was 66.6% (4/6), which was higher than that in rectal swabs (20.0%); however, the difference was not statistically significant. The detection rate of
K. septempunctata via PCR in the stool samples of 8 patients who consumed
P. olivaceus sashimi was 100.0% (4/4), which was significantly higher than that in the rectal swabs (25.0%) (
P=0.004). However, this study had some limitations. PCR test results obtained using each patient’s stool and rectal swab should be compared, but it was not possible to do so in practice.
One P. olivaceus sashimi sample that was confirmed to be positive for K. septempunctata was collected from a restaurant where a foodborne outbreak had occurred. To the best of our knowledge, this is the first study to identify K. septempunctata in both patients and food with epidemiological relevance in Korea.
Genotype of K. septempunctata
K. septempunctata is genetically classified into 3 groups (ST1, ST2, and ST3), and its genotype is determined by the combination of 2 mitochondrial genes,
cox1 and
rnl (
Table 2) [
11].
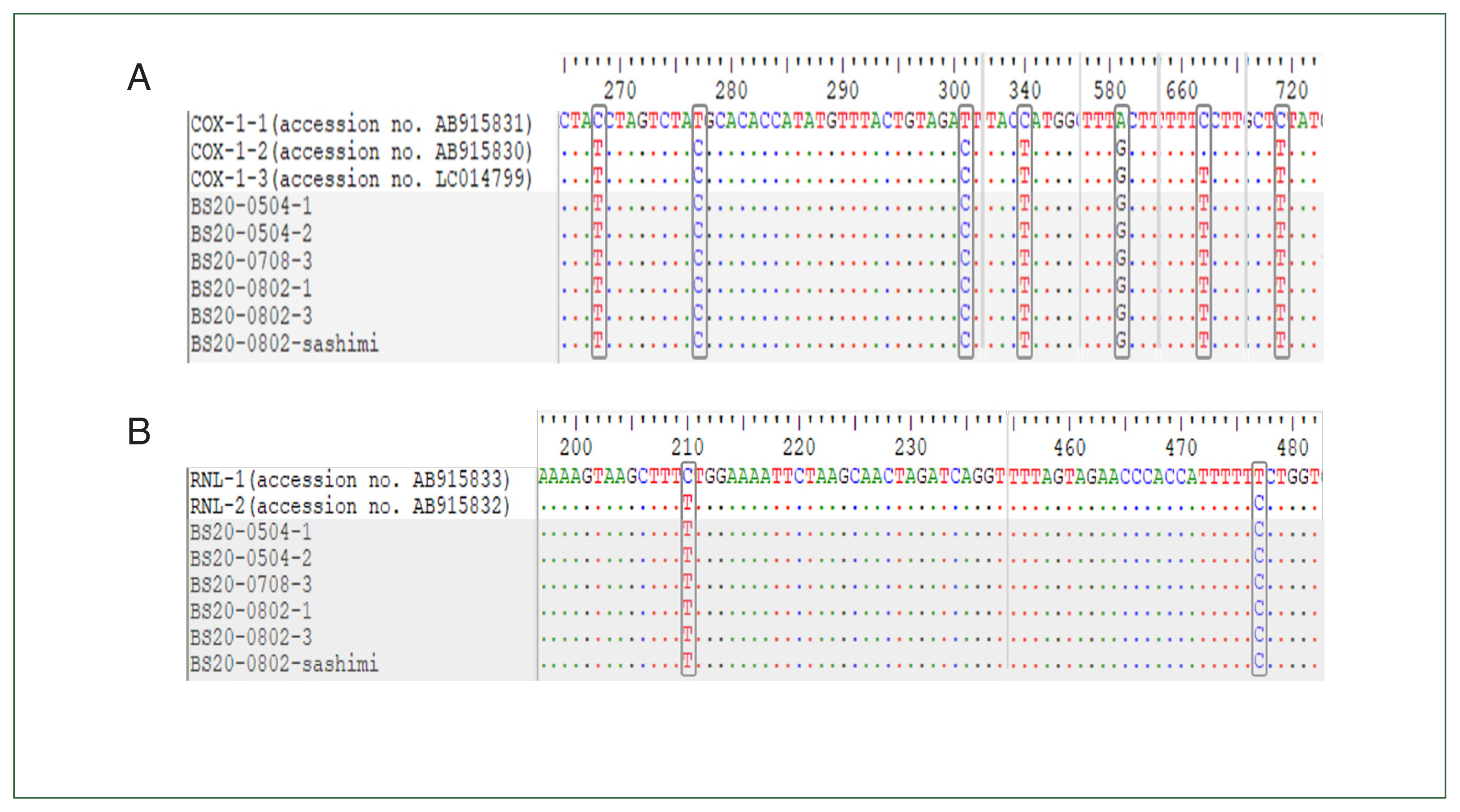
For the 5 samples collected from patients with foodborne illness (4 stool samples and 1 rectal swab sample) and 1 raw
P. olivaceus food sample,
cox1 and
rnl were subjected to multiple sequence alignment with sequences of standard strains. All samples were found to be of the ST3 genotype, comprising
cox1-3 and
rnl-2 (
Fig. 1).
Discussion
None of the 348 stool specimens collected from patients with acute diarrhea at the Enter-Net participating hospitals in Busan was found to contain
K. septempunctata. This finding was in contrast to the disease surveillance statistics of Enter-Net Korea, where detection rates of acute gastroenteritis-causing bacteria and viruses were 16.3 and 17.3%, respectively, during the year 2020 [
21]. The data obtained through epidemiological investigation reflect the following facts:
Kudoa food poisoning has a short incubation period; symptoms are relieved within 24 h; and the only causative food is
P. olivaceus sashimi infected with
K. septempunctata [
6,
18]. Similar results were obtained in the present study. The 5 patients who tested positive for
K. septempunctata presented a mean incubation time of 4 h and recovered within 3–20 h of symptom onset. The patient in outbreak C developed diarrhea and vomiting within 4 h of consuming
P. olivaceus sashimi and recovered after 11 h. The patient’s rectal swab sample was collected 58 h after ingestion of sashimi and no
K. septempunctata was detected. This finding was consistent with that of Kim et al. [
18], who suggested that epidemiological investigations should be conducted as early as possible because the detection rate of
K. septempunctata in feces was reported to fall rapidly after a time interval of 28.5 h from food intake to epidemiological survey.
Obtaining stool samples from patients with
Kudoa food poisoning is crucial. In this study, the detection rate of
K. septempunctata in the stool samples of 8 patients who consumed
P. olivaceus sashimi was 100.0%, which was significantly higher than that in rectal swab samples (25.0%). Kim et al. [
18] also reported a high detection rate of
K. septempunctata in stool samples (69.2%). Lee et al. [
19] reported that 31 (52.5%) of the 59 food poisoning outbreaks related to restaurants in Chungcheongnam-do, Korea, occurred at seafood restaurants; of these, 5 (8.5%) outbreaks were confirmed to be related to
Kudoa food poisoning. They tested for
Kudoa infection only in the stool among the collected fecal specimens and proposed that the detection rate of
K. septempunctata would have been higher if more number of stool samples had been collected. During epidemiological investigation of food poisoning outbreaks, in the absence of a tool to collect stools or if the patient fails to collect them, rectal swab specimens can be used instead of stool specimens. According to the Guidelines for the Management of Waterborne and Food Poisoning Infections [
22], rectal swabs are still used to detect bacterial and viral foodborne pathogens. However, only stool and vomit samples are recommended as diagnostic specimens for
Kudoa food poisoning. Therefore, few attempts have been made to detect
K. septempunctata in rectal swabs, even if
Kudoa infection is suspected due to the ingestion of raw fish, such as
P. olivaceus. This study is the first to genetically identify
K. septempunctata in one of the 5 (20.0%) rectal swab specimens. Similarly, a previous study attempted the molecular detection of enteric pathogens, including protozoa, in rectal swabs. A total of 15 pathogens (3 viruses, 3 parasites, and 9 bacteria) were identified using a commercial multiplex PCR assay. Several protozoal pathogens (
Giardia,
Cryptosporidium, and
Entamoeba histolytica) were identified in rectal swabs. The authors noted that flocked rectal swabs significantly facilitate the molecular diagnosis of diarrheal disease in children [
23]. The Annual Report of the Epidemiological Investigation of Infectious Diseases in Korea, published by the KDCA, reported 377 foodborne outbreaks in public restaurants nationwide in 2018, of which 211 (55.9%) had an unknown causative pathogen [
24]. However, the proportion of rectal swabs in the diagnostic specimens of these 211 foodborne outbreaks was not recorded. Nevertheless, implementing rectal swab testing for
K. septempunctata detection could dramatically improve the identification rate of
Kudoa infections. Therefore, although the detection rate of
K. septempunctata in rectal swab samples was lower than that in stool specimens, we strongly recommend the use of rectal swabs as diagnostic specimens for
Kudoa food poisoning.
K. septempunctata detected in a total of 6 specimens were all identified as the ST3 genotype. In a previous study by Takeuchi et al. [
11], ST1, ST2, and ST3 genotypes of
K. septempunctata were all reported to cause food poisoning. ST1 and ST2 genotypes are mainly detected in Japanese
P. olivaceus, whereas the ST3 genotype is detected in Korean
P. olivaceus. Moreover, the Korea Center for Disease Control and Prevention reported the presence of
K. septempunctata in 84 clinical specimens from 45 foodborne outbreaks that occurred in Korea from 2015 to autumn 2016. All strains were of the ST3 genotypes [
25], which is consistent with our results. To the best of our knowledge, this is the first study to detect
K. septempunctata in both patient and food specimens (
P. olivaceus sashimi) with epidemiological relevance in Korea. Previous animal experiments found that more than 10
6 Kudoa spores/mouse are needed to induce illness [
5]. However, in the present study, no microscopic examination was conducted to confirm the number of
Kudoa spores. Moreover, the
P. olivaceus sashimi samples collected were stored in restaurants where foodborne outbreaks had occurred and were not remnants from patient meals. Despite these limitations, the epidemiological relationship between these specimens cannot be ignored because they were collected within 24 h of food poisoning, and none of the 20 known food poisoning pathogens [
16] were identified (data not shown,
http://heis.busan.go.kr/health/virus_007_01.aspx).
In the present study, the samples were subjected to a genotyping method for
K. septempunctata based on mitochondrial (
cox1 and
rnl) gene polymorphisms. Moreover, we performed molecular epidemiological analysis of
K. septempunctata. Using PCR-restriction fragment length polymorphism (PCR-RFLP) analysis on partial mitochondrial DNA (mtDNA) encoding cytochrome b and NADH dehydrogenase subunit 1, Yokoyama et al. [
26] reported that
K. septempunctata is divided into 4 types. Ohnishi et al. [
27] identified 8 groups of
K. septempunctata by applying random amplified polymorphic DNA (RAPD) analysis. This genetic diversity suggests that additional genotypes exist in
K. septempunctata, which may impart different levels of pathogenicity in humans. Currently, there is no conclusive pathogenicity of ST3 in Korea [
28], and therefore further studies are required to identify more diverse genotypes for
K. septempunctata isolates.
In conclusion, we identified K. septempunctata 18S and 28S rDNA genes in 5 patients with food poisoning who ingested P. olivaceus sashimi and 1 sample of P. olivaceus sashimi. All detected K. septempunctata were genotype ST3, which has been commonly reported in Korea. P. olivaceus sashimi samples were collected from a restaurant visited by 2 patients in whom K. septempunctata genes were detected. This provides possible evidence that K. septempunctata is a causative pathogen for food poisoning. In addition, this study is the first to confirm the presence of K. septempunctata genes in a rectal swab sample. While suspected occurrences of Kudoa foodborne outbreaks continue, the identification rate of K. septempunctata remains low. One reason for this is the limitation in obtaining diagnostic specimens for Kudoa food poisoning, i.e., stool and vomit samples. Therefore, we strongly recommend obtaining rectal swabs as early as possible following symptom onset as diagnostic specimens for Kudoa food poisoning.
Notes
-
Author contributions
Conceptualization: Sung GH,Park EH
Data curation: Sung GH
Formal analysis: Sung GH
Investigation: Sung GH, Park IJ, Koo HS
Validation: Park EH, Lee MO
Writing – original draft: Sung GH
Writing – review & editing: Sung GH
-
The authors declare no potential conflicts of interest.
Fig. 1Comparison of nucleotide sequences among alleles of cox1 (A) and rnl (B) genes. Shaded area indicates the nucleotide sequence of 6 Kudoa septempunctata isolates from patients and food (sashimi).
Table 1Primers used for PCR amplification and sequencing
Table 1
|
Gene |
|
Primer sequence (5′-3′) |
Size (bp) |
Reference |
|
18S rDNA |
1st-F |
GGTGGGAGCATTTATTAGACT |
333 |
[26] |
|
1st-R |
AATCGAGACCACTGTCAAC |
|
2nd-F |
AGAAATACCGGAGTGGACCGTAAAATG |
|
2nd-R |
GTT CCA TGC TAT AAC ATT CAA GCG TTCG |
|
28S rDNA |
1st-F |
TGC GAG TGA AGC GGG AAA A |
356 |
|
1st-R |
GTG TTT CAA GAC GGG TCG G |
|
2nd-F |
GTG TGT GAT CAG ACT TGA TAT G |
|
2nd-R |
AAG CCA AAA CTG CTG GCC ATT T |
|
|
cox1
|
1st-F |
TTTGTTCATCGGCACAATTC- |
751 |
[9] |
|
1st-R |
ATAGCCTGGAACAAGGAATC |
|
2nd-F |
TATGGCAAAGAAGGTCTGAT |
500 |
|
2nd-R |
TCTAGGGATTCCACAAAGAC |
|
rnl
|
1st-F |
TGCCGTCAATTCTGTTGTATT |
817 |
|
1st-R |
AATACCCATGCTGTGTTCAT |
|
2nd-F |
GTTCCAACAAGTCCATGAA |
500 |
|
2nd-R |
GACTTTATGGACAACTCAGC |
Table 2
Kudoa septempunctata genotypes
Table 2
|
Genes (Accession no.) |
Genotypes |
|
|
ST1 |
ST2 |
ST3 |
|
cox1 1 (AB915831) |
+ |
− |
− |
|
cox1 2 (AB915830) |
− |
+ |
− |
|
cox1 3 (LC014799) |
− |
− |
+ |
|
|
rnl 1 (AB915833) |
+ |
− |
− |
|
rnl 2 (AB915832) |
− |
+ |
+ |
Table 3Summary of epidemiological characteristics for 11 food poisoning patients related to seafood intake in Busan
Table 3
|
Patient No. |
Outbreaks |
Date of occurrence |
Time interval (h)a
|
Incubation time (h)b
|
Diarrhea |
Vomiting |
Abdominal pain |
Intake of raw P. O. |
Sample type |
PCR result of Kudoac
|
|
1 |
BS20-0504-1 |
A |
05/04/20 |
38 |
4 |
+ |
+ |
+ |
Yes |
Stool |
D |
|
2 |
BS20-0504-2 |
A |
05/04/20 |
38 |
5 |
+ |
+ |
+ |
Yes |
Stool |
D |
|
3 |
BS20-0706-1 |
C |
07/06/20 |
58 |
4 |
+ |
+ |
− |
Yes |
Rectal swab |
ND |
|
4 |
BS20-0707-1 |
D |
07/07/20 |
20 |
5 |
+ |
− |
+ |
Yes |
Rectal swab |
ND |
|
5 |
BS20-0708-1 |
E |
07/08/20 |
16 |
3.5 |
+ |
+ |
+ |
Yes |
Rectal swab |
ND |
|
6 |
BS20-0708-3 |
E |
07/08/20 |
16 |
3 |
− |
− |
+ |
Yes |
Rectal swab |
D |
|
7 |
BS20-0802-1 |
F |
08/02/20 |
24 |
4 |
+ |
+ |
+ |
Yes |
Stool |
D |
|
8 |
BS20-0802-2 |
F |
08/02/20 |
24 |
4 |
+ |
+ |
− |
Yes |
Stool |
D |
|
9 |
BS20-0504-3 |
B |
05/04/20 |
25 |
6 |
+ |
− |
+ |
No |
Stool |
ND |
|
10 |
BS20-0504-4 |
B |
05/04/20 |
25 |
8 |
+ |
− |
+ |
No |
Stool |
ND |
|
11 |
BS20-0708-2 |
E |
07/08/20 |
16 |
5 |
− |
− |
+ |
No |
Rectal swab |
ND |
References
- 1. Moran J, Whitaker D, Kent M. A review of the myxosporean genus Kudoa Meglitsch, 1947, and its impact on the international aquaculture industry and commercial fisheries. Aquaculture 1999;172(1–2):163-196. http://doi.org/10.1016/S0044-8486(98)00437-2
- 2. Eiras JC, Saraiva A, Cruz C. Synopsis of the species of Kudoa Meglitsch, 1947 (Myxozoa: Myxosporea: Multivalvulida). Syst Parasitol 2014;87(2):153-180. http://doi.org/10.1007/s11230-013-9461-4
- 3. Yokoyama H, Whipps CM, Kent ML, Mizuno K, Kawakami H. Kudoa thyrsites from Japanese flounder and Kudoa lateolabracis n. sp. from Chinese sea bass: causative myxozoans of post-mortem myoliquefaction. Fish Pathol 2004;39(2):79-85. http://doi.org/10.3147/jsfp.39.79
- 4. Matsukane Y, Sato H, Tanaka S, Kamata Y, Sugita-Konishi Y. Kudoa septempunctata n. sp.(Myxosporea: Multivalvulida) from an aquacultured olive flounder (Paralichthys olivaceus) imported from Korea. Parasitol Res 2010;107(4):865-872. http://doi.org/10.1007/s00436-010-1941-8
- 5. Kawai T, Sekizuka T, Yahata Y, Kuroda M, Kumeda Y, et al. Identification of Kudoa septempunctata as the causative agent of novel food poisoning outbreaks in Japan by consumption of Paralichthys olivaceus in raw fish. Clin Infect Dis 2012;54(8):1046-1052. http://doi.org/10.1093/cid/cir1040
- 6. Sugita-Konishi Y, Sato H, Ohnishi T. Novel foodborne disease associated with consumption of raw fish, olive flounder (Paralichthys olivaceus). Food Saf 2014;2(4):141-150. http://doi.org/10.14252/foodsafetyfscj.2014026
- 7. Song JY, Choi JH, Choi HS, Jung SH, Park M. Monitoring of Kudoa septempunctata in cultured olive flounder and wild fish in Jeju Island during 2012. J Fish Pathol 2013;26(3):129-137. (in Korean). https://doi.org/10.7847/jfp.2013.26.3.129
- 8. Song JY, Kim MJ, Choi HS, Jung SH. Monitoring Kudoa septempunctata in cultured olive flounder Paralichthys olivaceus in different regions of Korea in 2013. Kor J Fish Aquat Sci 2014;47:611-621. (in Korean). http://doi.org/10.5657/KFAS.2014.0611
- 9. Food and Agriculture Organization. The State of World Fisheries and Aquaculture 2016. Contributiong to food security and nutrition for all. Food and Agriculture Organization of the United Nations; Rome, Italy. . 2016, pp 4-29.
- 10. Jung JY, Kim S, Kim K, Lee BJ, Kim KW, et al. Feed and disease at olive flounder (Paralichthys olivaceus) farms in Korea. Fishes 2020;5(3):21. http://doi.org/10.3390/fishes5030021
- 11. Takeuchi F, Ogasawara Y, Kato K, Sekizuka T, Nozaki T, et al. Genetic variants of Kudoa septempunctata (Myxozoa: Multivalvulida), a flounder parasite causing foodborne disease. J Fish Dis 2016;39(6):667-672. http://doi.org/10.1111/jfd.12395
- 12. Ahn M, Woo H, Kang B, Jang Y, Shin T. Effect of oral administration of Kudoa septempunctata genotype ST3 in adult BALB/c mice. Parasite 2015;22:35-39. http://doi.org/10.1051/parasite/2015035
- 13. Jang Y, Ahn M, Bang H, Kang B. Effects of Kudoa septempunctata genotype ST3 isolate from Korea on ddY suckling mice. Parasite 2016;23:18-24. http://doi.org/10.1051/parasite/2016020
- 14. Koo HS, Park JY, Sung GH, Park EH, Ku PT, et al. A study on Kudoa septempunctata infection from sashimi and sushi of olive flounder Paralichthys olivaceus in Busan, South Korea. Fish Aquat Sci 2021;24(8):277-283. http://doi.org/10.47853/FAS.2021.e27
- 15. Chung KS. Epidemiologic Investigation of Infectious Diseases in Korea Annual Report 2015. Korea Disease Control and Prevention Agency; Osong, Korea . 2016, pp 228-241.
- 16. Guideline for Water & Foodborne Diseases Prevention and Control 2017. Korea Disease Control and Prevention Agency; Osong, Korea. 2017, pp 46-337.
- 17. Lee SU. Analysis of Kudoa septempunctata as a cause of foodborne illness and its associated differential diagnosis. Epidemiol Health 2017;39:e2017014. http://doi.org/10.4178/epih.e2017014
- 18. Kim JJ, Ryu S, Lee H. Foodborne illness outbreaks in Gyeonggi province, Korea, following seafood consumption potentially caused by Kudoa septempunctata between 2015 and 2016. Osong Public Health Res Perspect 2018;9(2):66-72. http://doi.org/10.24171/j.phrp.2018.9.2.05
- 19. Lee H, Nam HS, Choi J, Park S, Park J, et al. Analysis of food poisoning outbreaks occurred in Chungnam Korea, 2019. J Environ Health Sci 2020;46(2):184-191. (in Korean). https://doi.org/10.5668/JEHS.2020.46.2.184
- 20. Ministry of Food and Drug Safety. Manual for Detection of Foodborne Pathogens at Outbreaks. Ministry of Food and Drug Safety; Osong, Korea. 2019, pp 313-320.
- 21. Korea Disease Control and Prevention Agency. Infectious disease statistics. Public Health Weekly Report 2021;14:83. (in Korean)..
- 22. Korea Disease Control and Prevention Agency. Guidelines for the Management of Waterborne and Food Poisoning Infections 2020. Korea Disease Control and Prevention Agency; Osong, Korea. 2020, pp 376-381.
- 23. Goldfarb DM, Steenhoff AP, Pernica JM, Chong S, Luinstra K, et al. Evaluation of anatomically designed flocked rectal swabs for molecular detection of enteric pathogens in children admitted to hospital with severe gastroenteritis in Botswana. J Clin Microbiol 2014;52(11):3922-3927. http://doi.org/10.1128/JCM.01894-14
- 24. Korea Disease Control and Prevention Agency. Epidemiological Investigation of Infectious Diseases in Korea Annual Report 2018. Korea Disease Control and Prevention Agency; Osong, Korea. 2019, p 17.
- 25. Cheun HI. Analysis on Foodborne Illness Cases of Kudoa septempunctata and Genotyping. Program and abstracts of the 58th annual meeting of the Korean society for parasitology and tropical medicine. . Korea: 2016; 21.
- 26. Yokoyama H, Mekata T, Satoh J, Nishioka T, Mori KI. Morphological and molecular comparisons between Japanese and Korean isolates of Kudoa septempunctata (Myxozoa: Multivalvulida) in the olive flounder Paralichthys olivaceus. Fish Pathol 2017;52(3):152-157. http://doi.org/10.3147/jsfp.52.152
- 27. Ohnishi T, Furusawa H, Oyama R, Koike S, Yoshinari T, et al. Molecular epidemiological analysis of Kudoa septempunctata by random amplified polymorphic DNA analysis. Jpn J Infect Dis 2015;68(3):235-238. http://doi.org/10.7883/yoken.JJID.2014.190
- 28. Chung YB, Bae JM. Is there evidence that Kudoa septempunctata can cause an outbreak of acute food poisoning? Epidemiol Health 2017;39:e2017004. http://doi.org/10.4178/epih.e2017004
- 29. Grabner D, Yokoyama H, Shirakashi S, Kinami R. Diagnostic PCR assays to detect and differentiate Kudoa septempunctata, K. thyrsites and K. lateolabracis (Myxozoa, Multivalvulida) in muscle tissue of olive flounder (Paralichthys olivaceus). Aquaculture 2012;338–341:36-40. http://doi.org/10.1016/j.aquaculture.2012.01.022