Abstract
Naegleria fowleri invades the brain and causes a fatal primary amoebic meningoencephalitis (PAM). Despite its high mortality rate of approximately 97%, an effective therapeutic drug for PAM has not been developed. Approaches with miltefosine, amphotericin B, and other antimicrobials have been clinically attempted to treat PAM, but their therapeutic efficacy remains unclear. The development of an effective and safe therapeutic drug for PAM is urgently needed. In this study, we investigated the anti-amoebic activity of Pinus densiflora leaf extract (PLE) against N. fowleri. PLE induced significant morphological changes in N. fowleri trophozoites, resulting in the death of the amoeba. The IC50 of PLE on N. fowleri was 62.3±0.95 μg/ml. Alternatively, PLE did not significantly affect the viability of the rat glial cell line C6. Transcriptome analysis revealed differentially expressed genes (DEGs) between PLE-treated and non-treated amoebae. A total of 5,846 DEGs were identified, of which 2,189 were upregulated, and 3,657 were downregulated in the PLE-treated amoebae. The DEGs were categorized into biological process (1,742 genes), cellular component (1,237 genes), and molecular function (846 genes) based on the gene ontology analysis, indicating that PLE may have dramatically altered the biological and cellular functions of the amoeba and contributed to their death. These results suggest that PLE has anti-N. fowleri activity and may be considered as a potential candidate for the development of therapeutic drugs for PAM. It may also be used as a supplement compound to enhance the therapeutic efficacy of drugs currently used to treat PAM.
-
Key words: Pinus densiflora, Naegleria fowleri, anti-amoebic activity, RNA sequencing, therapeutic drug candidate
Introduction
Naegleria fowleri is a free-living amoeba ubiquitously found in diverse natural environments, including fresh water and soil. However, it can infect humans and cause a fatal brain infection called primary amoebic meningoencephalitis (PAM) [
1]. The disease progresses very fast, resulting in death within 10 days after the onset of symptoms. Cases of PAM have been reported globally [
2], and concern for the expansion of this disease associated with climate change has increased in recent years [
3]. Although the global cases of PAM are rare, its high mortality rate (97%) emphasizes the urgency to develop effective therapeutics [
2,
4]. Treatment approaches involving miltefosine, amphotericin B, and other antimicrobial drugs have been attempted; however, their therapeutic efficacy is not guaranteed due to low clinical success and strong adverse effects [
5].
Natural products, including plants, have been widely used to manage or treat diverse diseases, including infectious diseases. Some of them have been applied as traditional medicine in different types or forms in many countries. Recently, attempts to discover natural compounds in diverse natural resources that have amoebicidal or ani-amoebic activity against pathogenic free-living amoebae have been performed [
6–
11]. These studies indicated the substantial amoebicidal or anti-amoebic activities of natural compounds or plant extracts against pathogenic amoeba species and their potential applications for therapeutic purposes or drug development for amoebic diseases.
Pinus densiflora (red pine), a member of the Pinaceae family, is widely distributed in East Asian countries, including Korea, China, Japan, and southeastern Russia [
12].
P. densiflora leaf (or needle) is used as food or traditional medicine in these countries because it contains diverse beneficial components that have a broad spectrum of pharmacological activities, such as antioxidant, antibacterial, antidiabetic, anti-inflammatory, anticancer, antihypertensive, and antithrombotic activities [
13–
19].
The aim of the present study was to evaluate the anti-amoebic activity of P. densiflora leaf extract (PLE) against N. fowleri trophozoites. PLE showed promising anti-amoebic activity against the trophozoites but low cytopathic effects in a glial cell line (C6). Furthermore, it induced subcellular morphological and physiological changes in the trophozoites, resulting in death. These findings suggest that PLE could be considered as a potential source for the development of a novel therapeutic drug or an alternative therapeutic strategy for PAM.
Materials and Methods
Cultivation of N. fowleri
Naegleria fowleri (Carter NF69 strain, ATCC 30215) was cultured in Nelson’s medium supplemented with 2% heat-inactivated fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% penicillin–streptomycin (P/S; Gibco) at 37°C. The amoeba was maintained by subculturing every 3 days, and all experiments were performed using trophozoites harvested during the early logarithmic growth phase.
Cultivation of glial cells
C6 glial cells (ATCC CCL-107) from rats were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% FBS (Gibco) and 1% P/S (Gibco). The cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
PLE preparation
Fresh leaves of
P. densiflora were collected during the spring (February to April) of 2018 at Goksung, Jeollanam-do, Korea, washed with distilled water several times, and dried. The juice of the leaves was extracted using a mechanical juicer (Inwha Precision and Samkwang, Gwangju, Korea) and freeze-dried in a vacuum freeze dryer (Samwon SFDSF12, Busan, Korea) at −70°C to −85°C. The collected leaf powder was dissolved in sterile distilled water, extracted at 60°C for 16 h, and freeze-dried. The resultant pellet was dissolved in sterile distilled water (1 g/ml) and filtered using a 0.2-μm syringe filter (Hyundai Micro, Seoul, Korea). The finally prepared water-soluble PLE was used for further experiments in this study (
Supplementary Fig. S1).
The viability of N. fowleri trophozoites and C6 cells after treatment with PLE was determined by a colorimetric method using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; VWR, Radnor, PA, USA). The amoeba trophozoites were seeded (105 cells/well) in 96-well plates (Thermo Fisher Scientific, Waltham, MA, USA) and incubated at 37°C for 6 h for cell attachment. The amoebae were treated with different concentrations of PLE (0–1,000 μg/ml) and incubated at 37°C for 48 h. The supernatant was drained from the plate at the indicated time points, and 20 μl of MTT (5 mg/ml) was added to each well, following which the plate was incubated at 37°C for 4 h. The supernatant was then aspirated, and 150 μl of dimethyl sulfoxide (DMSO; Sigma, St. Louis, MO, USA) was added to each well. The plate was incubated at 37°C for 30 min, and the reaction was read at 595 nm using a Multiskan FC microplate reader (Thermo Fisher Scientific). The C6 glial cells (2×104/well) were also seeded onto 96-well plates and treated with the same concentrations of PLE, following which the cell viability was assessed as described above.
Transmission electron microscopy (TEM)
Naegleria fowleri trophozoites treated with PLE (500 μg/ml) for 24 and 48 h were harvested, washed 3 times with phosphate-buffered saline (PBS, pH 7.4), and centrifuged at 200 g for 5 min. Amoeba cells not treated with PLE were also prepared using the same protocol and used as negative controls. The amoebae were fixed with 4% ice-cold glutaraldehyde and postfixed with 1% osmium tetroxide in 0.1 M phosphate buffer (pH 6.9) for 1 h. After fixation, the amoebae were washed in distilled water, dehydrated in an ascending series of ethanol concentrations, and embedded in an epoxy resin (Embed-812; Electron Microscopy Sciences, Hatfield, PA, USA). The blocks were sliced into ultrathin sections (90 nm), which were transferred onto 200-mesh copper grids (Agar Scientific, Stansted, UK) and stained with 4% uranyl acetate-lead citrate. The subcellular morphological structures of the amoebae were examined using a Talos L120C Cryo Bio 120 kV Transmission Electron Microscope (Thermo Fisher Scientific) at the Gyeongsang National University Center for Research Facilities (Jinju, Korea).
RNA sequencing (RNA-seq)
Total RNA was isolated from
N. fowleri trophozoites treated with PLE (500 μg/ml) for 24 h (
n=3) using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. Untreated amoebae (
n=3) were used as negative controls. The purified RNA was dissolved in RNase-free water, and the quality was determined by analyzing its purity and integrity using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The RNA concentration was determined using a DeNovix DS-11 spectrophotometer (Wilmington, DE, USA). A total amount of 1 μg of RNA (A260/A280 ratio >2.0; RNA integrity number >6.5) from each sample was used to construct the cDNA library. cDNA was synthesized using the QIAseq FX Single-cell RNA library kit (Qiagen, Valencia, CA, USA). The size and quality of each constructed library were confirmed using an Agilent 2100 high-sensitivity DNA chip (Agilent Technologies, Santa Clara, CA, USA). The library was amplified using a NovaSeq 6000 system (Illumina, San Diego, CA, USA) for 5-gigabase (Gb) in-depth sequencing. Trim Galore (
https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was used to assess the quality of the raw sequence reads, and those (base quality ≥30 min; length ≥50) available for further analyses were selected by read trimming.
Transcript quantification was performed by mapping all the preprocessed RNA-seq paired reads for each sample, and the finally assembled transcriptome was obtained using the Bowtie2 aligner (
https://bowtie-bio.sourceforge.net/bowtie2/index.html). The expression levels of mapped paired reads were normalized using Fragments Per Kilobase of transcript per Million (FPKM) mapped reads.
Semi-quantitative RT-PCR was performed to determine the expression profiles of the DEGs in the RNA-seq results. The amoebae treated with or without PLE were prepared as described above. Total RNA was isolated from the amoebae using the same method as described above. The RNA concentration in each sample was measured, equalized, and used for cDNA synthesis. A total of 3 μg of RNA was converted to cDNA using the RNA to cDNA EcoDry Premix (Clontech, Mountain View, CA, USA) according to the manufacturer’s protocols. Semi-quantitative RT-PCR was performed for the genes encoding
N. fowleri Kelch repeat protein (NF0105540), attractin-like protein 1 (ATRNL1; NF0044360), NADH azoreductase (NF0044710), and pyridoxine 5-phosphate oxidase (NF0073400);
N. fowleri glyceraldehyde-3-phosphate dehydrogenase (NfGAPDH) was amplified as an internal control [
21].
One-way analysis of variance (ANOVA) was performed on the normalized probes using P-values based on permutation (1,000 permutations per probe) with the overall threshold set at 0.05 (GraphPad Software, San Diego, CA, USA).
Results
PLE showed anti-amoebic activity against the N. fowleri trophozoites
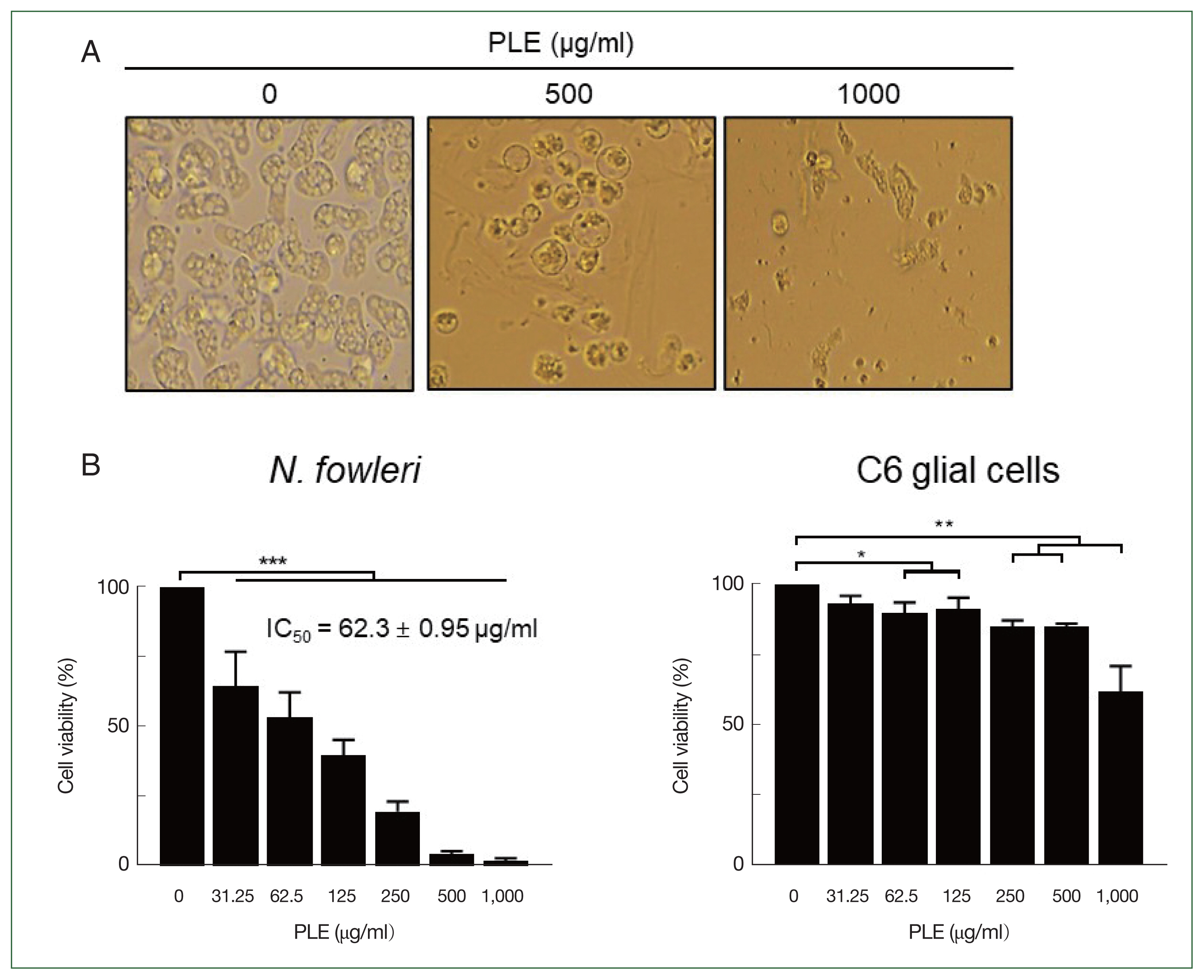
PLE showed anti-amoebic activity against the
N. fowleri trophozoites in a dose-dependent manner. It induced morphological changes, such as rounded shapes and size reductions in the amoebae (
Fig. 1A). A significant decrease in the viability of the amoebae was detected, with a half-maximal inhibitory concentration (IC
50) value of 62.3±0.95 μg/ml (
Fig. 1B). Alternatively, PLE did not cause any morphological changes of the C6 glial cells, although a partial cytotoxic effect was observed at high concentrations (
Fig. 1B). These results indicated the selective anti-amoebic activity of PLE against
N. fowleri trophozoites without causing any significant cytopathic effects on the C6 glial cells.
TEM analysis revealed that PLE induced ultrastructural morphological changes in the
N. fowleri trophozoites (
Fig. 2). Partial degradation of cellular organelles, reduction of contractile vacuoles, appearance of cellular compartments such as dense granules or blebs, and apoptotic bodies were observed in the PLE-treated amoebae. Additionally, an increase in abnormal mitochondria was observed.
RNA-seq analysis was performed to investigate the changes in the gene expression profiles of the PLE-treated (
n=3) and non-treated (
n=3)
N. fowleri. A total of 50.5 Gb, corresponding to 334,404,250 reads, was obtained from the samples. The number of total reads from each sample varied from 43,517,586 to 65,803,680. After quality control, 324,736,706 clean reads, differing by samples from 42,447,380 to 63,541,328, were selected. An assembly of clean reads produced 15,761 reliable contigs. The group comparisons revealed DEGs between the 2 groups, PLE-treated and non-treated amoebae (
P<0.05;
Fig. 3A). Compared to those in the PLE-untreated amoebae, 2,189 and 3,657 genes were significantly upregulated and downregulated, respectively, in the PLE-treated amoebae (
Fig. 3B). The sequences were mapped to GO terms and annotated, resulting in 3 clusters belonging to biological processes (1,742 genes), cellular components (1,237 genes), and molecular functions (846 genes;
Fig. 3C). Genes associated with binding, catalytic activity, and cellular structures were remarkably downregulated in the PLE-treated amoebae. Genes related to the regulation of biological processes and responses to stimuli were downregulated. Out of the 2,189 upregulated DEGs in the PLE-treated amoebae, the top 20 genes were succinate dehydrogenase, unspecific products, Kelch repeat protein, WD-40 repeat-containing protein, NADH azoreductase, alkane 1-monooxygenase, serine protease family, PX domain-containing protein kinase-like protein, attractin-like protein 1 (ATRNL1), predicted proteins, and NADP-dependent oxidoreductase (
Table 1). Alternatively, hemerythrin-like metal-binding protein, predicted proteins, dynein heavy chain, nucleolar basal body binding protein bn46 51 small subunit, unspecified products, PB1 domain-containing protein, transforming protein, ankyrin repeat domain protein, synaptobrevin vesicle-associated membrane protein (VAMP), dynein gamma flagellar outer arm, membrane protein, hypothetical protein, and pyridoxine 5-phosphate oxidase were the top 20 downregulated genes (
Table 1).
The expression profiles of 4 DEGs in the PLE-treated and non-treated
N. fowleri trophozoites were comparatively analyzed by semi-quantitative RT-PCR to evaluate the quality of the RNA-seq analysis. Consistent with the RNA-seq results, the genes showed different expression levels between the 2 amoebae groups (
Fig. 4).
Discussion
A promising anti-amoebic activity of PLE against
N. fowleri was observed in this study. PLE induced remarkable morphological changes in the
N. fowleri trophozoites, resulting in the death of the amoebae. However, it exhibited no cytotoxic effects against the C6 glial cells. Transcriptome analysis via RNA-seq was performed to understand the molecular mechanism involved in the anti-amoebic activity of PLE. Remarkable changes in the expression levels of diverse genes were detected in the PLE-treated amoebae. A total of 5,846 DEGs (2,189 upregulated and 3,657 downregulated) were identified. GO analysis revealed that the expression levels of genes associated with molecular function and cellular components were greatly suppressed in the PLE-treated amoebae. Genes related to essential biological processes, such as regulation of biological processes and response to stimuli, were also markedly downregulated in the amoebae. However, most genes upregulated and downregulated in the PLE-treated amoebae were unidentified products, predicted proteins, or hypothetical proteins. Out of 176 genes that were upregulated by more than 4-folds in the PLE-treated amoebae compared to those in the non-treated amoebae, 68 (38.6%) were unspecified products, predicted proteins, and hypothetical proteins; likewise, among the 686 genes that were downregulated by >4-folds, 255 (37.2%) were unidentified products, predicted proteins, and hypothetical proteins. Considering the highly restricted biological information of
N. fowleri proteins, it is difficult to predict the molecular functions or pathways involved in the anti-amoebic effect of PLE against
N. fowleri. However, the enhanced expression levels of genes encoding proteins potentially associated with protein-protein interactions and signal pathways, such as Kelch repeat protein, WD-40 repeat-containing protein, PX domain-containing protein kinase-like protein, ATRNL1, and ankyrin repeat domain protein [
22–
26], indicate that massive cellular and physiological alterations that are not observed under normal conditions in the PLE-treated amoebae. The expression of enzymes associated with cellular defense against stressed conditions, such as NADH azoreductase and alkane 1-monooxygenase [
26,
27], was also significantly increased in PLE-treated
N. fowleri. Conversely, the expression levels of genes encoding enzymes that participate in metabolic functions, cellular motility, and protein interactions, such as hemerythrin-like metal-binding protein, dynein heavy chain, nucleolar basal body binding protein bn46 51 small subunit, and PB1 domain-containing protein [
28–
32], were significantly downregulated in the PLE-treated amoebae. In addition to these genes, the overall expression levels of genes essential for cellular homeostasis, protein synthesis, energy production, and cellular motility, such as transcription factors, ribosomal proteins, vacuolar proteins, and actin-associated proteins, were also decreased in the PLE-treated
N. fowleri trophozoites. These findings suggest that PLE induces dramatic changes in the biological and cellular functions of
N. fowleri, which may eventually contribute to death. However, further comprehensive studies are necessary to determine the underlying molecular mechanisms involved in the anti-amoebic activity of PLE against
N. fowleri.
In conclusion, this study demonstrated the anti-amoebic activity of PLE against N. fowleri. PLE caused morphological and biological changes in N. fowleri trophozoites, but did not exhibit any significant cytotoxicity against glial cells. PLE may be used as an alternative therapy or for the development of anti-PAM drugs. However, further isolation and characterization of active compounds with anti-amoebic activity in PLE are necessary. Additionally, in vivo studies evaluating the therapeutic effects of PLE or PLE-derived compounds against PAM are warranted.
Notes
-
The authors declare no conflict of interest.
-
Conceptualization: Na BK
Data curation: Lê HG
Formal analysis: Lê HG, Kang JM, Võ TC, Yoo WG, Na BK
Funding acquisition: Na BK
Investigation: Kang JM, Na BK
Methodology: Lê HG, Kim W, Kang JM, Võ TC, Cheong H
Project administration: Na BK
Resources: Kim W, Cheong H
Software: Lê HG
Supervision: Na BK
Validation: Lê HG, Na BK
Visualization: Lê HG
Writing – original draft: Lê HG, Na BK
Writing – review & editing: Kim W, Kang JM, Võ TC, Yoo WG, Cheong H
Supplementary Information
Acknowledgments
This research was supported by a National Research Foundation of Korea (NRF) grant from the Government of Korea (NRF-2021R1A2C1091855).
Fig. 1Anti-amoebic activity of PLE on N. fowleri trophozoites. (A) Morphological change. Treatment with PLE (500 or 1,000 μg/ml) for 48 h induced significant morphological changes in N. fowleri trophozoites. (B) Viability. Different concentrations of PLE (0 to 1,000 μg/ml) were added to N. fowleri trophozoites or C6 cells and incubated for 48 h. PLE caused a remarkable reduction in the viability of N. fowleri trophozoites with an IC50 value of 62.33±0.95 μg/ml. However, it did not significantly affect the viability of the C6 glial cells. The viability of the amoebae and C6 cells is presented as a percentage relative to the negative controls (not treated with PLE). The results are presented as the mean and standard deviation (error bar) from 3 independent assays. *P<0.01, **P<0.001, ***P<0.0001.
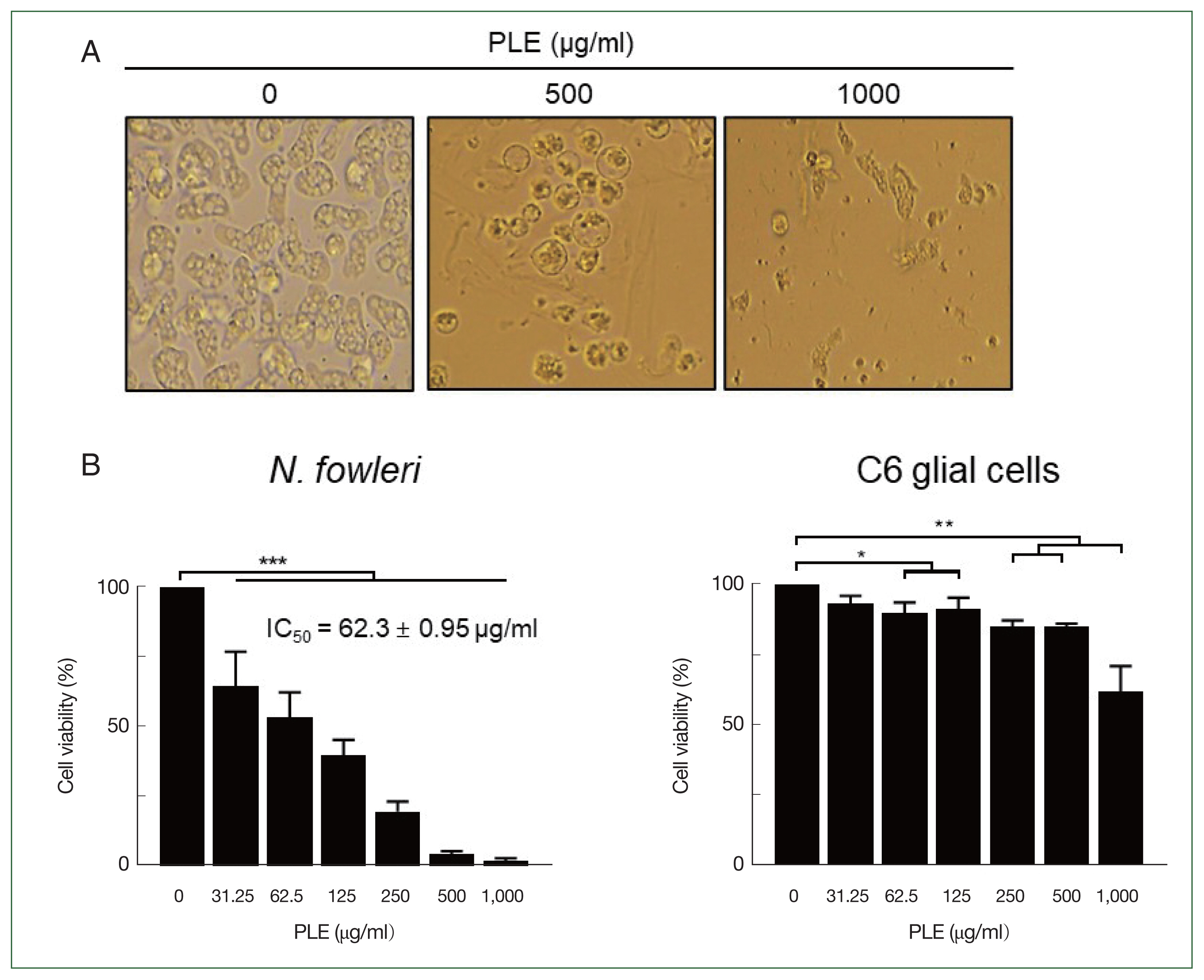
Fig. 2Transmission electron microscopy (TEM) analysis of N. fowleri trophozoites treated with PLE. Naeglaria fowleri trophozoites treated with PLE (500 μg/ml) for 24 h and 48 h showed remarkable ultrastructural morphological changes. Red circles indicate putative apoptotic bodies. Blebs (blue arrowheads). Autophagy-like vacuoles (yellow arrowheads). Magnification, 6,000× or 10,000×. CV, contractile vacuoles; M, mitochondria; N, nucleus.
Fig. 3DEGs in the PLE-treated and non-treated N. fowleri. (A) MA plot for the DEGs. The red crosses represent significantly upregulated or downregulated genes, and the black spots indicate the non-DEGs. Pink spots represent DEGs, but they were not statistically significant. (B) DEG expression pattern. RNA-seq analysis revealed 5,846 DEGs (2,189 upregulated and 3,657 downregulated) in the PLE-treated N. fowleri. (C) Gene ontology (GO) analysis. The top 11 significantly enriched GO terms of the target genes were related to biological processes, cellular components, and molecular function.

Fig. 4Semi-quantitative RT-PCR. The expression levels of genes encoding N. fowleri Kelch repeat protein (NF0105540), attractin-like protein 1 (ATRNL1; NF0044360), NADH azoreductase (NF0044710), and pyridoxine 5-phosphate oxidase (NF0073400) were comparatively analyzed in the PLE-treated and non-treated N. fowleri. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified as an internal control.
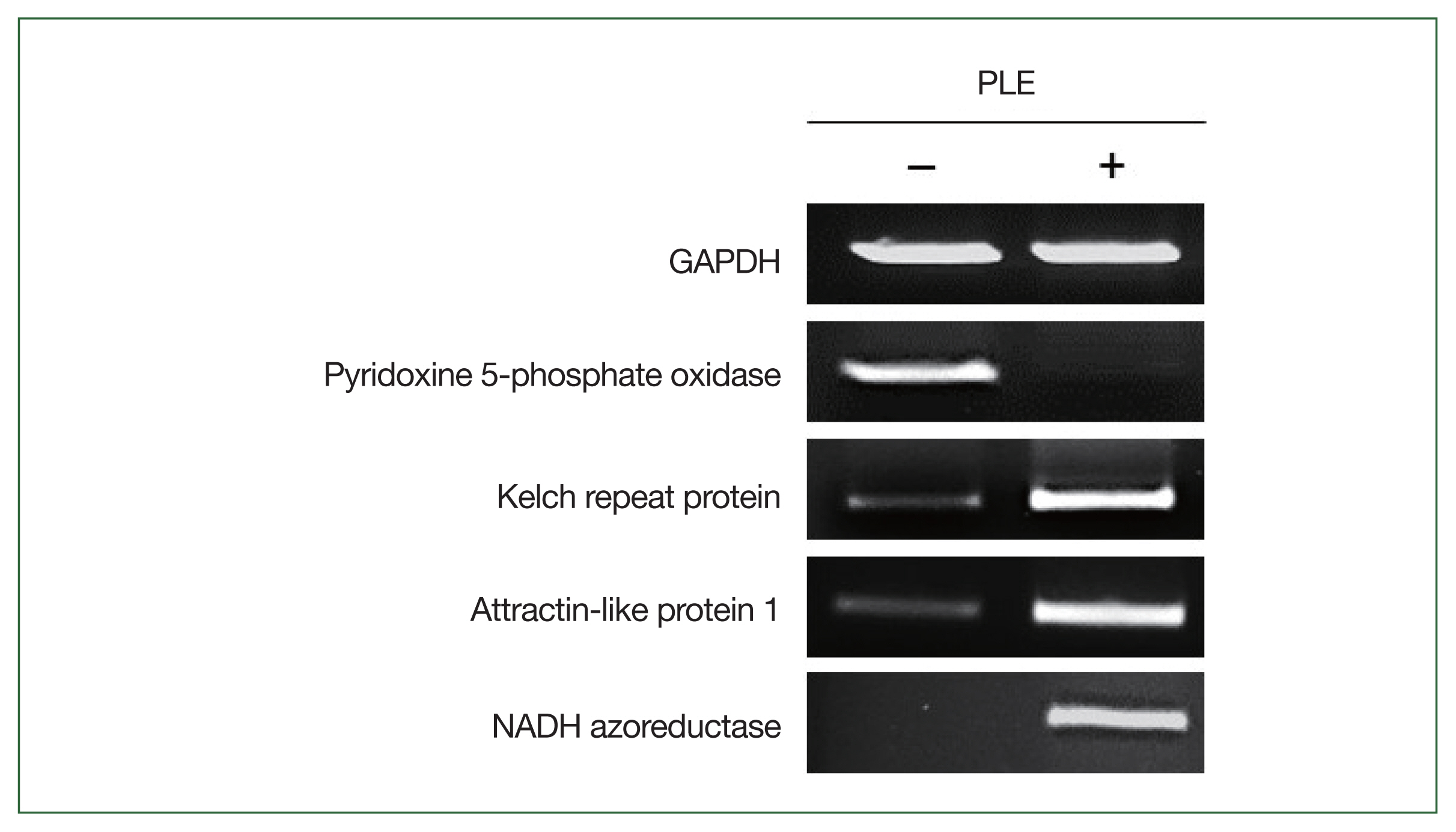
Table 1Top 20 up-regulated and down-regulated DEGs in PLE-treated N. fowleri
Table 1
|
Gene |
Gene ID |
log2 (FC) |
|
Up-regulation |
|
Succinate dehydrogenase |
NF0040010 |
8.58 |
|
Unspecified product |
NF0132120 |
7.31 |
|
Unspecified product |
NF0114330 |
7.11 |
|
Unspecified product |
NF0041760 |
7.05 |
|
Unspecified product |
NF0004850 |
6.74 |
|
Kelch repeat protein |
NF0105540 |
6.56 |
|
WD-40 repeat-containing protein |
NF0076760 |
6.40 |
|
NADH azoreductase |
NF0044710 |
6.40 |
|
Alkane 1-monooxygenase |
NF0077100 |
6.35 |
|
Serine protease family |
NF0106810 |
6.29 |
|
PX domain-containing protein kinase-like protein |
NF0059050 |
6.25 |
|
Unspecified product |
NF0015450 |
6.08 |
|
Unspecified product |
NF0002010 |
6.05 |
|
Attractin-like protein 1 (ATRNL1) |
NF0044360 |
6.05 |
|
Predicted protein |
NF0007650 |
5.95 |
|
Unspecified product |
NF0043100 |
5.91 |
|
Predicted protein |
NF0011170 |
5.82 |
|
NADP-dependent oxidoreductase |
NF0072900 |
5.64 |
|
Unspecified product |
NF0045970 |
5.61 |
|
Unspecified product |
NF0127400 |
5.41 |
|
|
Down-regulation |
|
Hemerythrin-like metal-binding protein |
NF0127030 |
−9.79 |
|
Predicted protein |
NF0115530 |
−8.32 |
|
Dynein heavy chain |
NF0014710 |
−7.97 |
|
Nucleolar basal body binding protein bn46 51 small subunit |
NF0104690 |
−7.87 |
|
Unspecified product |
NF0013060 |
−7.59 |
|
Unspecified product |
NF0121520 |
−7.40 |
|
Unspecified product |
NF0092320 |
−7.30 |
|
PB1 domain containing protein |
NF0081890 |
−7.28 |
|
Transforming protein |
NF0112230 |
−7.28 |
|
Ankyrin repeat domain protein |
NF0110610 |
−7.19 |
|
Predicted protein |
NF0107550 |
−7.06 |
|
Synaptobrevin vesicle-associated membrane protein (VAMP) |
NF0078380 |
−7.03 |
|
PB1 domain containing protein |
NF0050000 |
−6.99 |
|
Predicted protein |
NF0079120 |
−6.93 |
|
Dynein gamma flagellar outer arm |
NF0014700 |
−6.89 |
|
Predicted protein |
NF0061820 |
−6.84 |
|
Membrane protein |
NF0031290 |
−6.84 |
|
Predicted protein |
NF0034030 |
−6.78 |
|
Hypothetical protein |
NF0059600 |
−6.75 |
|
Pyridoxine 5-phosphate oxidase |
NF0073400 |
−6.66 |
|
Unspecified product |
NF0035330 |
−6.66 |
References
- 1. Schuster FL, Visvesvara GS. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int J Parasitol 2004;34(9):1001-1027. https://doi.org/10.1016/j.ijpara.2004.06.004
- 2. Gharpure R, Bliton J, Goodman A, Ali IKM, Yoder J, et al. Epidemiology and clinical characteristics of primary amebic meningoencephalitis caused by Naegleria fowleri: a global review. Clin Infect Dis 2021;73(1):e19-e27. https://doi.org/10.1093/cid/ciaa520
- 3. Gharpure R, Gleason M, Salah Z, Blackstock AJ, Hess-Homeier D, et al. Geographic range of recreational water-associated primary amebic meningoencephalitis, United States, 1978–2018. Emerg Infect Dis 2021;27(1):271-274. https://doi.org/10.3201/eid2701.202119
- 4. Baig AM, Khan NA. Novel chemotherapeutic strategies in the management of primary amoebic meningoencephalitis due to Naegleria fowleri. CNS Neurosci Ther 2014;20(3):289-290. https://doi.org/10.1111/cns.12225
- 5. Haston JC, Cope JR. Amebic encephalitis and meningoencephalitis: an update on epidemiology, diagnostic methods, and treatment. Curr Opin Infect Dis 2023;36(3):186-191. https://doi.org/10.1097/QCO.0000000000000923
- 6. Boonhok R, Sangkanu S, Norouzi R, Siyadatpanah A, Mirzaei F, et al. Amoebicidal activity of Cassia angustifolia extract and its effect on Acanthamoeba triangularis autophagy-related gene expression at the transcriptional level. Parasitology 2021;148(9):1074-1082. https://doi.org/10.1017/S0031182021000718
- 7. Mitsuwan W, Sangkanu S, Romyasamit C, Kaewjai C, Jimoh TO, et al. Curcuma longa rhizome extract and curcumin reduce the adhesion of Acanthamoeba triangularis trophozoites and cysts in polystyrene plastic surface and contact lens. Int J Parasitol Drugs Drug Resist 2020;14:218-229. https://doi.org/10.1016/j.ijpddr.2020.11.001
- 8. Belofsky G, Carreno R, Goswick SM, John DT. Activity of isoflavans of Dalea aurea (Fabaceae) against the opportunistic ameba Naegleria fowleri. Planta Med 2006;72(2):383-386. https://doi.org/10.1055/s-2005-9162529
- 9. Lê HG, Choi JS, Hwang BS, Jeong YT, Kang JM, et al. Phragmites australis (Cav.) Trin. ex Steud. extract induces apoptosis-like programmed cell death in Acanthamoeba castellanii trophozoites. Plants 2022;11(24):3459. https://doi.org/10.3390/plants11243459
- 10. Siddiqui R, Boghossian A, Khatoon B, Kawish M, Alharbi AM, et al. Anti-amoebic properties of metabolites against Naegleria fowleri and Balamuthia mandrillaris. Antibiotics 2022;11(5):539. https://doi.org/10.3390/antibiotics11050539
- 11. Lê HG, Kang JM, Võ TC, Na BK. Kaempferol induces programmed cell death in Naegleria fowleri. Phytomedicine 2023;119:154994. https://doi.org/10.1016/j.phymed.2023.154994
- 12. Choung HL, Hong SK. Distribution patterns, floristic differentation and succession of Pinus densiflora forest in South Korea: a perspective at nation-wide scale. Phytocoenologia 2006;36(2):213229. https://doi.org/10.1127/0340-269X/2006/0036-0213
- 13. Jung MJ, Chung HY, Choi JH, Choi JS. Antioxidant principles from the needles of red pine, Pinus densiflora. Phytother Res 2003;17(9):1064-1068. https://doi.org/10.1002/ptr.1302
- 14. Süntar I, Tumen I, Ustün O, Keleş H, Küpeli Akkol E. Appraisal on the wound healing and anti-inflammatory activities of the essential oils obtained from the cones and needles of Pinus species by in vivo and in vitro experimental models. J Ethnopharmacol 2012;139(2):533-540. https://doi.org/10.1016/j.jep.2011.11.045
- 15. Wu Y, Bai J, Zhong K, Huang Y, Qi H, Jiang Y, et al. Antibacterial activity and membrane-disruptive mechanism of 3-p-trans-coumaroyl-2-hydroxyquinic acid, a novel phenolic compound from pine needles of Cedrus deodara, against Staphylococcus aureus. Molecules 2016;21(8):1084. https://doi.org/10.3390/molecules21081084
- 16. Jo JR, Park JS, Park YK, Chae YZ, Lee GH, et al. Pinus densiflora leaf essential oil induces apoptosis via ROS generation and activation of caspases in YD-8 human oral cancer cells. Int J Oncol 2012;40(4):1238-1245. https://doi.org/10.3892/ijo.2011.1263
- 17. Sang JL, Ki WL, Haeng JH, Ji YC, Seo YK, et al. Phenolic phytochemicals derived from red pine (Pinus densiflora) inhibit the invasion and migration of SK-Hep-1 human hepatocellular carcinoma cells. Ann N Y Acad Sci 2007;1095:536-544. https://doi.org/10.1196/annals.1397.058
- 18. Park J, Kim WJ, Kim W, Park C, Choi CY, et al. Antihypertensive effects of dehydroabietic and 4-epi-trans-communic acid isolated from Pinus densiflora. J Med Food 2021;24(1):50-58. https://doi.org/10.1089/jmf.2020.4797
- 19. Park J, Lee B, Choi H, Kim W, Kim HJ, et al. Antithrombosis activity of protocatechuic and shikimic acids from functional plant Pinus densiflora Sieb. et Zucc needles. J Nat Med 2016;70(3):492-501. https://doi.org/10.1007/s11418-015-0956-y
- 20. Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010;26(1):136-138. https://doi.org/10.1093/bioinformatics/btp612
- 21. Lê HG, Ham AJ, Kang JM, Võ TC, Naw H, et al. A novel cysteine protease inhibitor of Naegleria fowleri that is specifically expressed during encystation and at mature cysts. Pathogens 2021;10(4):388. https://doi.org/10.3390/pathogens10040388
- 22. Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol 2000;10(1):17-24. https://doi.org/10.1016/s0962-8924(99)01673-6
- 23. Stirnimann CU, Petsalaki E, Russell RB, Müller CW. WD40 proteins propel cellular networks. Trends Biochem Sci 2010;35(10):565-574. https://doi.org/10.1016/j.tibs.2010.04.003
- 24. Leonard TA, Hurley JH. Regulation of protein kinases by lipids. Curr Opin Struct Biol 2011;21(6):785-791. https://doi.org/10.1016/j.sbi.2011.07.006
- 25. Mosavi LK, Cammett TJ, Desrosiers DC, Peng Z. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci 2004;13(6):14351448. https://doi.org/10.1110/ps.03554604
- 26. Stark Z, Bruno DL, Mountford H, Lockhart PJ, Amor DJ. De novo 325 kb microdeletion in chromosome band 10q25.3 including ATRNL1 in a boy with cognitive impairment, autism and dysmorphic features. Eur J Med Genet 2010;53(5):337-339. https://doi.org/10.1016/j.ejmg.2010.07.009
- 27. Miller EN, Jarboe LR, Yomano LP, York SW, Shanmugam KT, et al. Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl Environ Microbiol 2009;75(13):4315-4323. https://doi.org/10.1128/AEM.00567-09
- 28. Karlsson CMG, Cerro-Gálvez E, Lundin D, Karlsson C, Vila-Costa M, et al. Direct effects of organic pollutants on the growth and gene expression of the baltic sea model bacterium Rheinheimera sp. BAL341. Microb Biotechnol 2019;12(5):892-906. https://doi.org/10.1111/1751-7915.13441
- 29. Alvarez-Carreño C, Alva V, Becerra A, Lazcano A. Structure, function and evolution of the hemerythrin-like domain superfamily. Protein Sci 2018;27(4):848-860. https://doi.org/10.1002/pro.3374
- 30. Asai DJ, Wilkes DE. The dynein heavy chain family. J Eukaryot Microbiol 2004;51(1):23-29. https://doi.org/10.1111/j.1550-7408.2004.tb00157.x
- 31. Sumimoto H, Kamakura S, Ito T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE 2007;2007(401):re6. https://doi.org/10.1126/stke.4012007re6
- 32. Trimbur GM, Walsh CJ. BN46/51, a new nucleolar protein, binds to the basal body region in Naegleria gruberi flagellates. J Cell Sci 1992;103(1):167-181. https://doi.org/10.1242/jcs.103.1.167
 , Woong Kim3,4, Jung-Mi Kang1, Tuấn Cường Võ1,2, Won Gi Yoo1,2
, Woong Kim3,4, Jung-Mi Kang1, Tuấn Cường Võ1,2, Won Gi Yoo1,2 , Hyeonsook Cheong3, Byoung-Kuk Na1,2,*
, Hyeonsook Cheong3, Byoung-Kuk Na1,2,*