Abstract
Free-living amoebae (FLA) are found in diverse environments, such as soils, rivers, and seas. Hence, they can be used as bioindicators to assess the water quality based solely on their presence. In this study, we determined the presence of FLA in river water by filtering water samples collected from various sites and culturing the resulting filtrates. FLA were detected in all the water samples with varying quality grades (Grades I–V). The significant increase in the size of the amoebae population with the deterioration in the water quality. Monoxenic cultures of the amoebae were performed, and genomic DNAs were isolated, among which 18S rDNAs were sequenced to identify the amoeba species. Of the 12 species identified, 10 belonged to the Acanthamoeba genus; of the remaining 2 species, one was identified as Vannella croatica and the other as a species of Vermamoeba. Acanthamoeba was detected in samples with Grades I to VI quality, whereas the Vermamoeba species was present only in Grade I water. V. croatica was found exclusively in water with Grade II quality. Following morphological observations, genomic DNA was sequenced using 16S rDNA to determine whether the species of Acanthamoeba harbored endosymbionts. Most of the isolated Acanthamoeba contained endosymbionts, among which 4 species of endogenous bacteria were identified and examined using transmission electron microscopy. This study provides evidence that the distribution of amoebae other than Acanthamoeba may be associated with water quality. However, further confirmation will be required based on accurate water quality ratings and assessments using a more diverse range of FLA.
-
Key words: Acanthamoeba, Vannella, Vermamoeba, environmental bioindicator, free living amoeba
Introduction
Bioindicators are organisms that indicate the quality of their environment [
1]. Thus, the environmental conditions of an area can be inferred by examining the status of these bioindicators. Inaccurate results may be obtained when performing direct measurements of the water quality due to factors such as the condition of the water and the weather on the day of measurement; the use of bioindicators can contribute to eliminating such inaccuracies [
2].
In Korea, the quality of water has been classified into 7 grades based on the Korean-Comprehensive Water Quality Index (or in terms of the biochemical oxygen demand): Grade Ia, wherein the water is considered suitable for domestic use with simple purification, and aquatic organisms such as scuds and crayfish can thrive in water of this quality; Grade Ib, wherein the water is considered suitable for domestic use following standard purification; Grade II, wherein the water can be used for swimming after standard purification (
Semisulcospira and
Plecoglossus are found in waters with these 2 grades); Grade V water is deemed normal and can be used for industrial purposes following standard purification (
Zacco platypus and specied in the genus
Pseudorasbora can be found in aquatic environments with water of this quality); Grade IV water is considered fairly poor and can be used as industrial water after having undergone advanced purification (species such as
Misgurnus spp. and
Silurus asotus are found in environments with water of this standard); Grade V is indicative of high levels of pollution, and these waters tend to be populated with more tolerant invertebrate species, such as those in the family Chironomidae and
Tubifex tubifex; and Grade VI, which has very poor quality and is unsuitable for most fish species [
3].
FLA tend to occur ubiquitously in a diverse range of terrestrial and aquatic habitats [
4], and some species, such as testate amoebae, have been used as bioindicators that are highly responsive to changes in environmental conditions. Species of testate amoebae are found in diverse environments, and their presence or absence in a particular environment can facilitate the rapid detection of environmental change. The dependence of these amoebae on specific ecosystems can indicate the health and stability of a particular ecosystem, thereby serving as a valuable diversity-based monitoring system [
5–
7].
In Korea, species of FLA in the genera
Acanthamoeba,
Vannella,
Naegleria, and
Vermamoeba are readily found in freshwater habitats [
8–
11] and are of particular medical importance. For example, the
Acanthamoeba species, which can exist as cysts or trophozoites, have been identified as causal agents of amoebic keratitis and granulomatous amoebic encephalitis [
12]. Alternatively, cysts of the
Vannella species have a fan-shaped morphology, and the trophozoites can change to a stellate form [
13,
14]. The
Naegleria species, which inhabit both soil and water, are ameboflagellate [
15] and can occur in one of 3 forms: cyst, trophozoite, and flagellate.
Naegleria fowleri has been identified as the causal organism of primary amoebic meningoencephalitis [
16].
Vermamoeba vermiformis, characterized by small round double-walled cysts and worm-like trophozoites, causes corneal inflammation and has also been reported to be associated with certain pathogenic bacteria, including
Legionella pneumophila [
17].
The aim of this study was to determine the potential use of FLA commonly found in Korean rivers as bioindicators by collecting water samples from aquatic habitats and determining the types and amounts of isolated amoebae.
Materials and Methods
Water samples
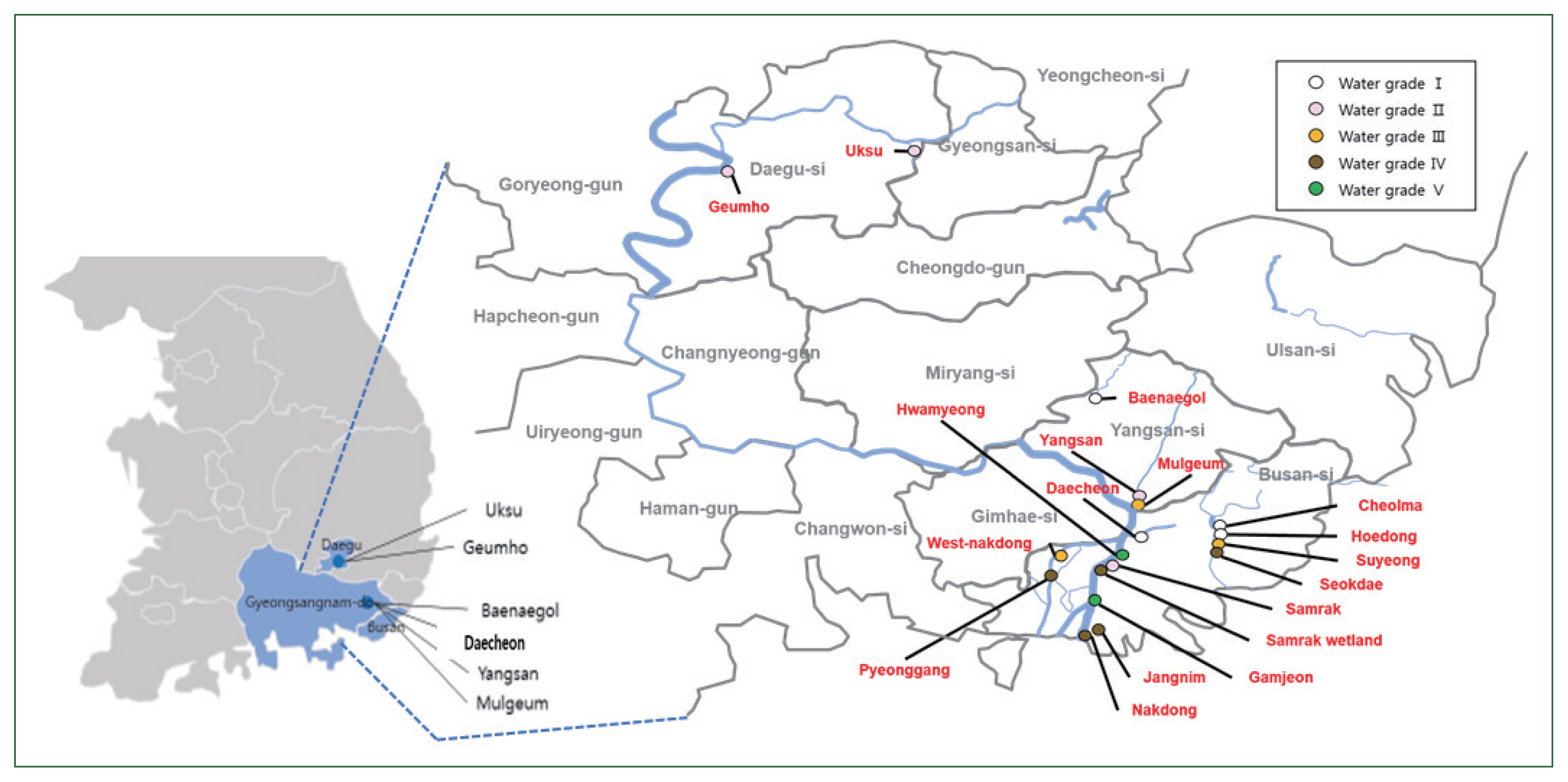
Water samples from multiple sites along the Nakdong River in Korea were collected in sterilized 1-L bottles (Winners, Busan, Korea). Each site was characterized by a specific water grade (
Fig. 1): Grade I (Ia and Ib)–Daecheon, Cheolma, Hoedong, Baenaegol, and Yusan; Grade II–Geumho, Samnak, Yangsan, and Uksu; Grade III–Mulgeum, Seonakdong, and Suyeong; Grade IV–Pyeonggang, Seokdae, Jangnim, Nakdong, and Samrak (wetland); and Grade V (V and VI)–Hwamyung and Gamjeon [
18,
19].
Agar medium for culturing the amoebae was prepared by initially autoclaving a 2% suspension of Bacto-agar (Biosciences, San Jose, CA, USA), which was cooled to a temperature of approximately 50°C–60°C and supplemented with amphotericin B solution (Sigma, St. Louis, MO, USA) to inhibit fungal growth. Agar plates were prepared by pouring 12 ml aliquots of the agar into Petri dishes (SPL, Pocheon, Korea); the plates were smeared with a 20% suspension of heated
Escherichia coli [
20].
Filtering apparatus (filter flask, glass base, graduated funnel, and aluminum clamp) that had initially been autoclaved and sterilized were used to filter the collected water samples. The water samples were filtered through 0.45-μm pore filters (Advantec membrane filters, Tokyo, Japan). The filters were lifted from the device using a scalpel, and forceps were inverted and placed on the surfaces of the E. coli-inoculated agar plates, which were then incubated at 25°C. The filtering apparatus was resterilized between the filtering of each sample to prevent cross-contamination.
Monoxenic and axenic cultures
Fungal growth and amoebae were microscopically checked every day after incubation. The amoebae were isolated, if detected. Regions on plates with dense amoebal growth were marked with a pen, excised, and placed on the surfaces of fresh agar plates. After obtaining the monoxenic cultures, we excised gel segments with good amoebal growth and placed them in a solution of 1% HCl, followed by incubation overnight at 25°C for axenization. The following day, after centrifuging at 1,500 rpm for 5 min, the resulting supernatants were removed, and 7 ml of peptone yeast extract glucose medium was added to the remaining pellets; the suspensions were then transferred to T-25 flasks, followed by incubation at 25°C [
20].
Genomic DNA was extracted from the cultured amoeba to identify the species of FLA and endogenous bacteria. The lysis buffer (SNET buffer) used to extract the DNA contained 20 mM Tris-HCl (pH 8.0, 1 M Tris-HCl; Dongin Biotech, Korea), 5 mM EDTA (pH 8.0, 0.5 M EDTA; Dongin Biotech), 400 mM NaCl (Duksan Pure Chemical, Ansan, Korea), 1% SDS (BiosesangTM, Yongin, Korea), and distilled water (DW), which was filtered before use using a 50-ml syringe and a 0.45-μm syringe filter (Sartorius Stedim Biotech, Göttingen, Germany). After extracting the amoebae from the 1.5% agar gels using 1× PBS, the suspensions were centrifuged at 15,000 rpm for 10 min. Following the removal of the supernatants, the remaining pellets were resuspended in protease K-supplemented SNET buffer at a concentration of 0.1 g/4 ml. The samples were incubated overnight in a 55°C-incubator using a rocker. The following day, an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) reagent was added to the samples, followed by incubation on a rocker for 30 min at room temperature. The mixtures were centrifuged at 15,000 rpm for 5 min at room temperature, and the resulting supernatants were transferred to fresh Eppendorf tubes. An equal volume of isopropanol was added, and the samples were centrifuged at 8,000 rpm for 15 min at 4°C. The resulting supernatants were discarded, and the pellets were washed with 1 ml of 70% ethanol by centrifuging at 8,000 rpm for 5 min at 4°C. The ethanol was discarded, following which the pellets were air-dried for 20 min and dissolved in diluted 1×TE buffer (10×TE buffer; Dongin Biotech).
Genetic characterization
Polymerase chain reaction (PCR) was performed to identify the amoebae isolates using reaction systems containing 2 μg of template DNA, 0.5 μl of Taq buffer, 3 μl of 10× buffer, 2 μl of dNTPs, and 2 μl each of the forward and reverse 18S rDNA primers (V4-1F 5′-GCGGTAATTCCAGCTC-3′ and V4-4R 5′-GCCMTTCCGTCAATTCC-3′) [
21]. The amplifications were performed with forward and reverse 16s rDNA primers (27F 5′-AGAGTTTGATCCTGGCTCAG-3′ and 1492R 5′-GGTTACCTTGTTACGACTT-3′) to identify the bacterial endosymbiont, and the total volume was adjusted to 30 μl using DW. The PCR conditions were as follows: initial denaturation at 95°C for 5 min, followed by 34 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 45 sec, and a final extension step at 72°C for 5 min, after which the samples were held at 4°C. The amplified products obtained were loaded onto 1% agar gels and subsequently sequenced commercially by Macrogene (Seoul, Korea).
The nucleotide sequences were compared with those of published strains via BLAST searches. Clustal X was used for pairwise alignment and to calculate the percent sequence identities. The percentage similarity corresponds to the number of identical sites between 2 isolates aligned against each other, as in the master alignment. The phylogeny was determined using the Clustal W2 program (
http://www.ebi.ac.uk/Tools/clustalw2/index.html) with a low gap penalty, and phylogenetic trees were constructed using TreeView software. All sequences obtained in this study have been registered in GenBank.
Samples were prepared for transmission electron microscopy by initially prefixing with 2.5% glutaraldehyde at 4°C, in a phosphate buffer (pH 7.2) and postfixing with 1% osmium tetroxide using the same buffer. The material was then dehydrated through a graded series of ethanol and embedded in epoxy resin (Epon 812 mixture). Sections (1 μm thick) were stained with 1% toluidine blue for examination under a light microscope. Thin sections (50–60 nm) were prepared using an ultramicrotome (EM UC7; Leica) and double-stained with uranyl acetate and lead citrate. The stained sections were observed under a JEM-1200EXII transmission electron microscope (JEOL).
Results
Initial detection of FLA on agar plates
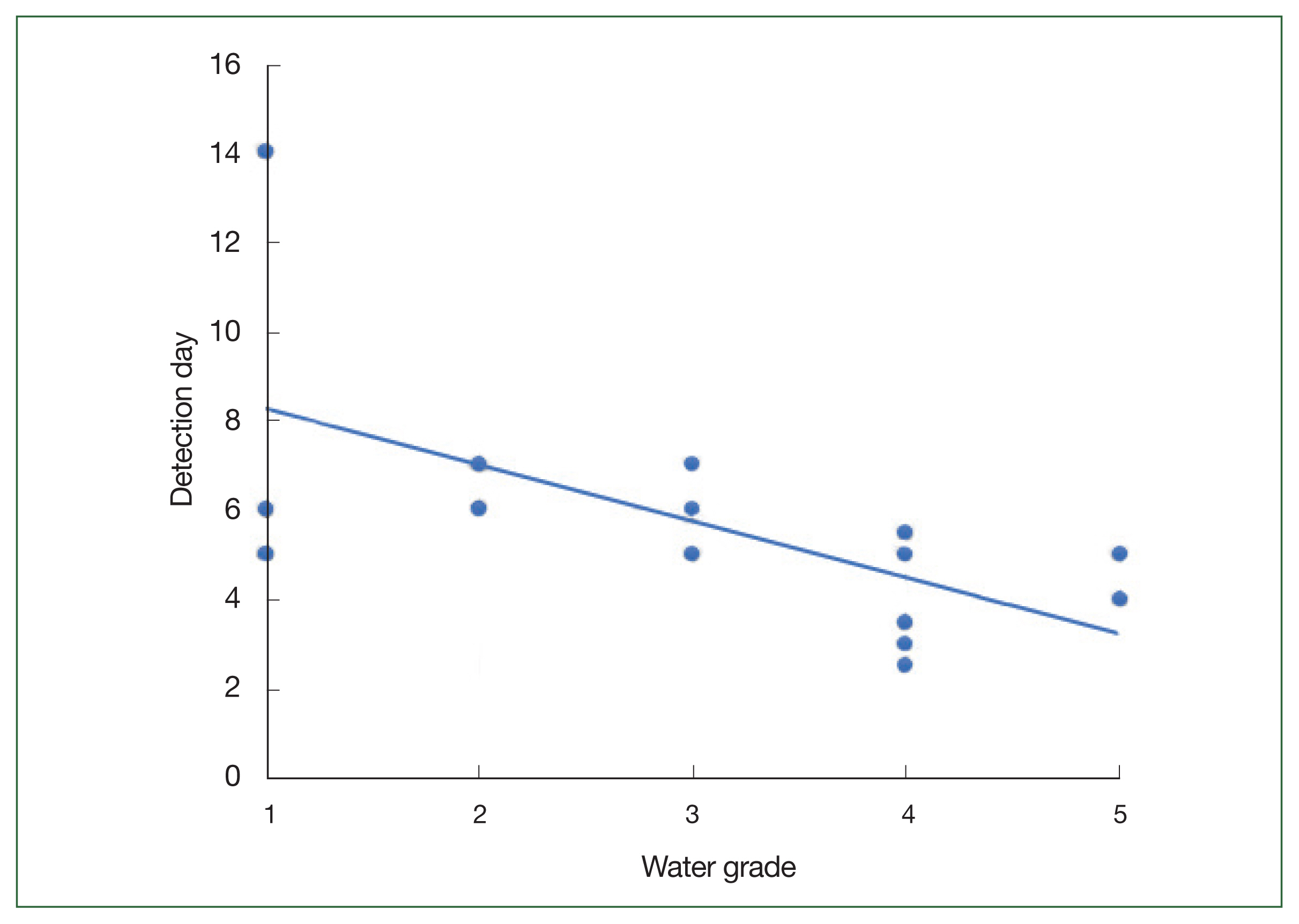
The filters through which 500 ml of fresh water had been passed were observed at daily intervals under a light microscope to determine differences in the populations of FLA based on the water quality. The time to initial detection of FLA decreased with the deterioration in the quality of the collected water sample (
Fig. 2). In the case of the Grade I water samples, FLA were initially observed over a protracted period of 5–14 days after inoculation, whereas in the Grade II and III samples, the FLA were detected within a week; the FLA were detected within 2 days in the Grade IV and V water samples. These observations indicated the increase in the level of FLA contamination as the quality of water deteriorated. Thus, amoeba in water samples of poor quality can be detected more rapidly when cultured on agar plates.
FLA were detected in all 18 water samples collected and cultured under monogenic and axenic conditions in this study. FLA were readily detected and isolated in most Grade I and II water samples. However, in poorer quality water (Grades III–V), the detection and isolation of the amoeba proved to be more difficult, owing to the rapid and prolific growth of bacteria and fungi. Consequently, only 3 FLA were isolated from the Grade III to V quality water samples (
Table 1). Two types of FLA were detected in samples collected from Cheolma and Hoedong, and monoxenic cultures of amoeba were obtained for all Grade II water samples, except those collected from Baenaegol and Yusan. In total, 4 species of amoebae were isolated in the genus
Acanthamoeba and one in the genus
Vermamoeba (
Hartmannella) from the Grade I water samples (KA/DC, KA/CM1, KA/CM2, KA/HD1, and KA/HD2). We obtained monoxenic cultures for all isolates detected in the Grade II water samples based on which 4 types of FLA (KA/GH, KA/SR, KA/YS, and KA/US) were identified; however, not all were successfully cultured in axenic medium. Three of these were identified as species of
Acanthamoeba, whereas the remaining isolate (KA/YS) was identified as
Vannella croatica, via molecular analysis. All 3 isolates obtained from the Grade III to V water samples were identified as species of
Acanthamoeba (KA/MG, KAHM, and KAGJ).
For the morphological analyses, a microscope was used to take photographs of amoebae derived from monogenic cultures at ×400 magnification (
Fig. 3). Among the identified isolates, the cysts of KA/DC appeared as small spheres, but no trophozoites were detected. KA/YS (a species of
Vannella), presented as notably large trophozoites (approximately 34 μm in length) that were not readily distinguishable from the cysts of this amoeba. Similarly, KA/GJ, a group I species of
Acanthamoeba, was characterized by large trophozoites (approximately 30 μm in length), whereas the morphology of the cysts in the KA/MG isolate (a group III species of
Acanthamoeba) differed notably from that of the cysts of other group II species of
Acanthamoeba. The remaining isolates, all group II species of
Acanthamoeba, consisted of both ecto- and endotype cysts (
Supplementary Fig. S1).
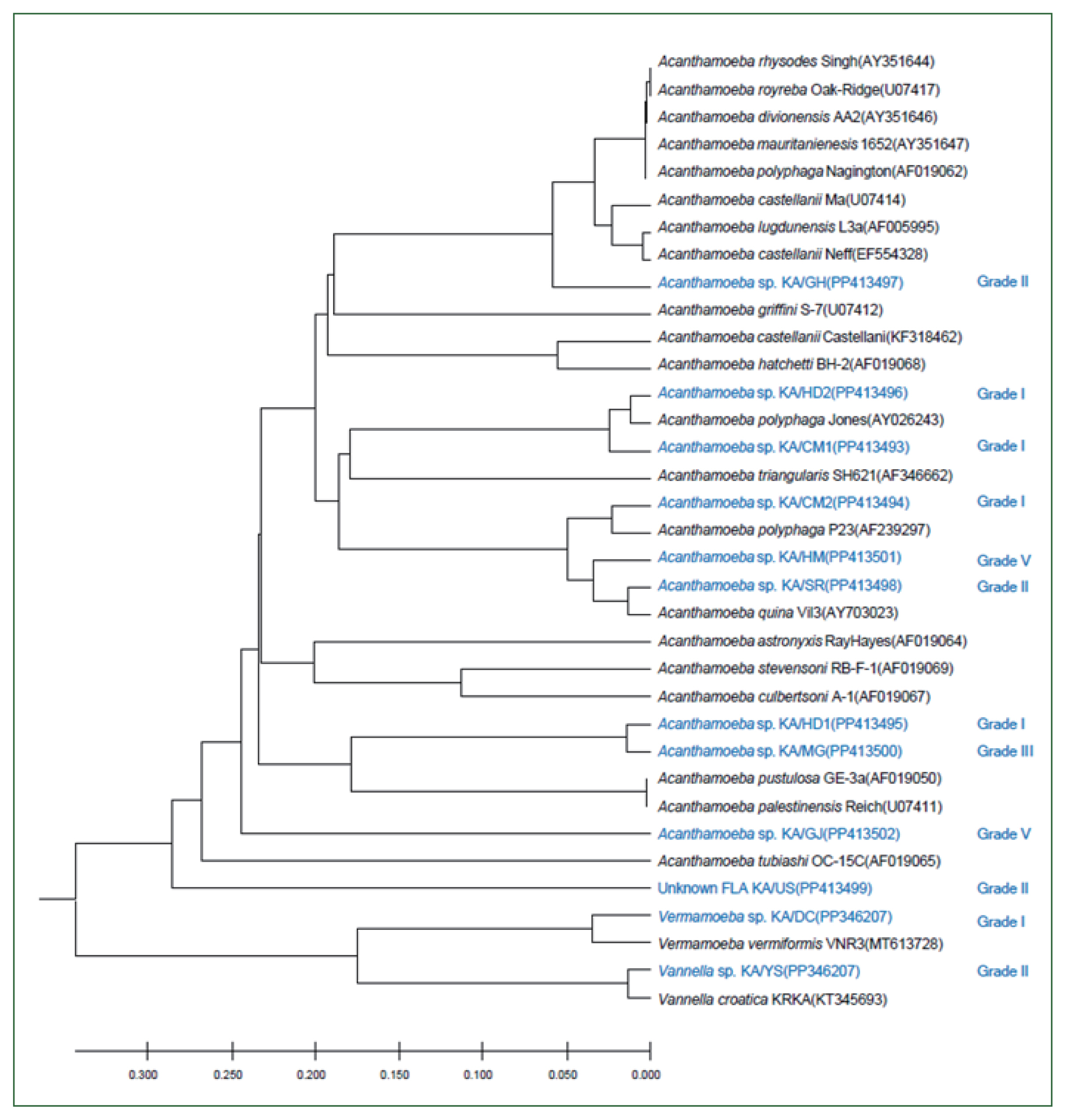
The 18S rDNA sequences of the 12 isolates were compared with those of the reference strains of 21
Acanthamoeba, one
Vermamoeba, and one
Vannella species using ClustalW. The genetic characteristics of most of these isolates were similar to those of the group II
Acanthamoeba species, with the sequences of KA/HD2, KA/CM1, and KA/CM2 being similar to those of type
A. polyphaga and
A. triangularis; alternatively, KA/GH appeared to be closely related to
A. castellani Neff and species in the
A. lugdunensis L3a complex. The KA/HM5 and KA/SR sequences were similar to that of
A. quina. The genetic distance between KA/HD1 and KA/MG was less than 0.01%, placing these isolates in the same group as the group III
Acanthamoeba species
A. pustulosa and
A. palesteinsis. The KA/DC isolate was closely related to species in the
Vermamoeba genus, and KA/YS was closely related to
Vannella croatica. In the case of KA/US, the sequence was genetically similar to the
Acanthamoeba species; however, a large genetic distance was observed between this isolate and most of
Acanthamoeba. Moreover, the KA/US sequence did not exhibit a close match with any of the reference strains; hence, it could be a previously unrecorded FLA (
Fig. 3).
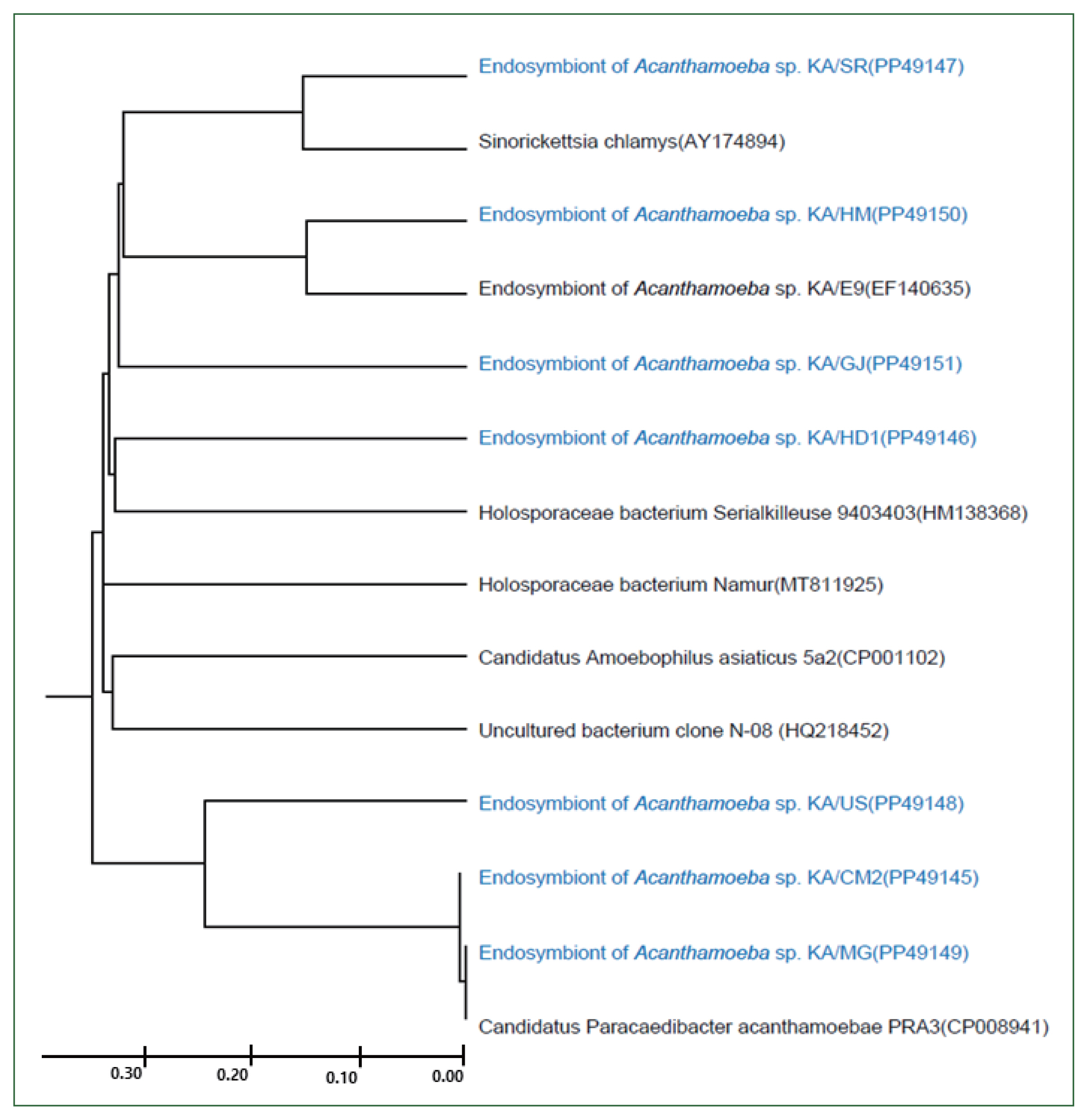
In this study, 8 FLA isolates were successfully cultured in axenic medium, and endosymbionts based on 16S rDNA PCR analysis, for which similar sequences have been reported previously (
Fig. 4), were detected in all of them, except for KA/CM1. KA/HD1 showed the highest similarity (97%) to the recently published 16S rDNA sequence of a
Holosporaceae bacterium Serialkilleuse 9,403,403 strain. The 16S rDNA sequences of the endosymbionts of KA/SR and KA/HM were found to be closely related to
Sinorichettsia chlamys and an endosymbiont of
Acanthamoeba sp. KA/E9, respectively. The putative endosymbionts in KA/US, KA/CM2, and KA/MG were closely related, with sequences similar to those of the Candidatus
Paracaedibacter acanthamoebae PRA3 strain.
The morphological and ultrastructural characters of the detected endosymbionts were elucidated via transmission electron microscopy (
Fig. 5). All the rod-shaped endosymbionts detected in the FLA isolates were estimated to be approximately 1.72 (1.70–1.75)×0.27 (0.20–0.30) μm in size (
Fig. 5A–E). In each case, the bacterial membrane was in close contact with the cytoplasm of the host FLA isolates. In isolate KA/SR, we detected a further bacteria-like structure within the cytoplasm, which lacked a membrane structure and appeared to be surrounded by a food vesicle (red arrowhead,
Fig. 5F). Although 16S rDNA was detected in the KA/GJ isolate, we could not identify any endosymbiont-like structure with the cytoplasm of these amoebae.
Discussion
The sustainability of FLA as bioindicators of water quality was assessed in this study. FLA from freshwater habitats characterized by varying qualities of water, ranging from Grade I to VI, were collected. As anticipated, FLA were detected in all the collected freshwater samples. Although we could not detect any significant differences among the species of FLA isolates based on the quality of the source water, the time to detection tended to shorten with the deterioration in water quality from Grade I to V. This phenomenon may indicate the higher nutrient content in more polluted water, which supports the larger populations of FLA [
22].
As shown in
Table 1, the number of coliform bacteria in the water samples increased with the decline in water quality. The poor-quality water may serve as a source of food for FLA inhabiting these habitats. The method used in the current study could serve as an additional approach for determining the water quality in different types of streams and rivers. The numbers of coliform bacteria were differences when comparing the water quality grades each time a sample collection (data not shown), indicating that numerous different approaches based on different criteria can be used to determine the water quality [
2].
A notable finding of this study is that, although FLA were detected in all the collected water samples, we isolated only 12 distinct types (
Table 1). In poor quality water (≥Grade III), obtaining monoxenic and axenic cultures of FLA is particularly difficult because of the heavy fungal and bacterial contamination. Thus, it is difficult to ascertain whether the inability to isolate FLA from Grade III and IV water reflected the effect of water quality on the presence of amoebae in this study. Nevertheless, among the FLA isolates, we discovered a species of
Vannella croatica, which, to the best of our knowledge, has not been reported from riverine waters in Korea. The genetic characteristics of this isolate (KA/YS) indicate that it is related to
Vannella croatica, the morphology and phylogenetic characteristics of which were first reported in Croatia in 2016 [
23]. However, only the trophozoite form of this species was detected; no cysts were observed in the current study. The size of the trophozoites ranged from 30 to 50 μm, and their movement was notably more rapid than that of the
Acanthamoeba species.
Microscopic observations of the FLA morphology were conducted, and the 18S rDNA genes were sequenced from the genomic DNA to identify the FLA at the species level [
4,
20]. The majority of the isolated FLA belonged to species in the genus
Acanthamoeba. Morphological analysis indicated that they were group I- to III-type
Acanthamoeba, with most having group II characteristics (
Supplementary Fig. S1). However, given that the shape of
Acanthamoeba can vary depending on the culture conditions, morphology-based assignments can prove unreliable [
4]. In the present study, amoeba with genetic characteristics similar to those of
Vermamoeba vermiformis were isolated from water samples of Grade I quality. Lee et al. [
24] indicated that, in Korea, 12.9% of tap water from buildings with storage tanks was contaminated with FLA, and the rate of contamination in highway service areas was as high as 33.3% [
24]. Nearly all the FLA detected by the authors were genetically similar to
Vermamoeba vermiformis (previously classified as
Hartmannella vermiformis) [
24]. The KFA5 isolate could represent a new allergen; the airway resistance values for this isolate were significantly elevated after 6 intranasal treatments, similar to values obtained following the administration of
Acanthamoeba [
25].
Furthermore, 16S rDNA PCR analyses of all axenic cultured isolates, except for the KA/CM1 isolate, were performed to determine the existence of endogenous bacteria in FLA. Endogenous bacteria were detected in all FLA, among which 4 species (Holosporaceae bacterium, Sinorickettsia chlamys, Candidatus Amoebophilus asiaticus, and Candidatus Paracaedibacter acanthamoebae) were identified. Based on these findings, it was impossible to discern any clear association between the endogenous bacteria in FLA and the quality of water from which the amoebae were isolated. Although genetically distinct, certain amoeba isolates (KA/US, KA/CM2, and KA/MG) hosted a similar complement of symbiotic bacteria. In contrast, genetically similar FLA (KA/HD1 and KA/MG) contained notably different symbiotic bacteria. Interestingly, although large numbers of Acanthamoeba previously isolated from tap water have been shown to lack bacterial endosymbionts, almost all amoebae isolated from river water were found to harbor symbiotic bacteria in this study. These findings indicate that the endosymbiotic associations between Acanthamoeba and bacteria can appear and disappear. In the present study, endosymbiotic bacteria were detected in the KA/GJ isolate based on the 16S rDNA PCR analysis but were not visible under the electron microscope.
Species in the
Holosporales order, which are an alphaproteobacterial lineage encompassing bacteria obligatorily associated with multiple diverse eukaryotes within isolated FLA [
26], were also detected in the current study. These bacteria are phylogenetically related to the symbionts of other ciliates and diplonemids, forming a putatively rapidly evolving clade within the
Holosporaceae family; the characteristic features of this clade include the presence of specific secretion systems, and they are speculated to have a mild parasitic effect on their hosts [
26]. The endosymbiont identified in the
Acanthamoeba KA/SR was found to be very similar to that detected in KA/E9 isolated from humans with keratitis [
27]. However, there is no strong evidence to indicate that this endosymbiont plays a pathogenic role. Interestingly,
Sinorickettsia chlamys (GenBank No. AY174894), previously identified as an intracellular proteobacterium in the Chinese scallop
Chlamys farreri, a species of marine bivalve, was identified in the
Acanthamoeba isolate KA/SR in this study. These bacteria are phagocytosed by different aquatic animals and subsequently persist as intracellular “organelles.” However, the roles played by these bacteria in animal cells have not been established and warrant further research.
WQI is a simple tool that assigns a single value to water quality by considering a certain number of biological, chemical, and physical parameters (also called variables) to represent the water quality in an easy and understandable way [
28,
29]. Despite the efforts of researchers around the world, an index that can be universally applied by water agencies, users, and managers in different countries is lacking [
30]. In the current study, we aimed to use FLA as an indicator of water quality. However, it takes a long time to isolate and culture amoeba, and using them with the currently available methods is challenging. Therefore, it is necessary to develop a simple kit by developing the PCR and Loop-Mediated Isothermal Amplication methods that can easily detect FLA in water samples.
In conclusion, although we were unable to provide conclusive evidence indicating an association between FLA (or their endosymbionts) and the quality of water from which they were isolated, we did detect significant increases in the sizes of the amoeba populations with the deterioration in the water quality. This result showed that eutrophication of fresh water is positively related to the proliferation of free-living amoeba.
Notes
-
Conceptualization: Choi A, Seong JW, Kim JH, Lee JY, Cho HJ, Yu HS
Data curation: Choi A, Seong JW, Kim JH, Lee JY, Cho HJ, Jeong MJ, Choi SY, Jeong YJ
Formal analysis: Jeong MJ
Investigation: Park MK, Jeong MJ
Methodology: Kang SA, Jeong MJ, Jeong YJ
Resources: Kang SA, Jeong YJ
Software: Kang SA
Supervision: Kang SA, Park MK
Writing – original draft: Choi A, Yu HS
Writing – review & editing: Yu HS
-
The authors declare that they have no conflict of interest.
Supplementary Information
Fig. 1Map showing the locations of the sampling sites. Water quality is indicated by the color.
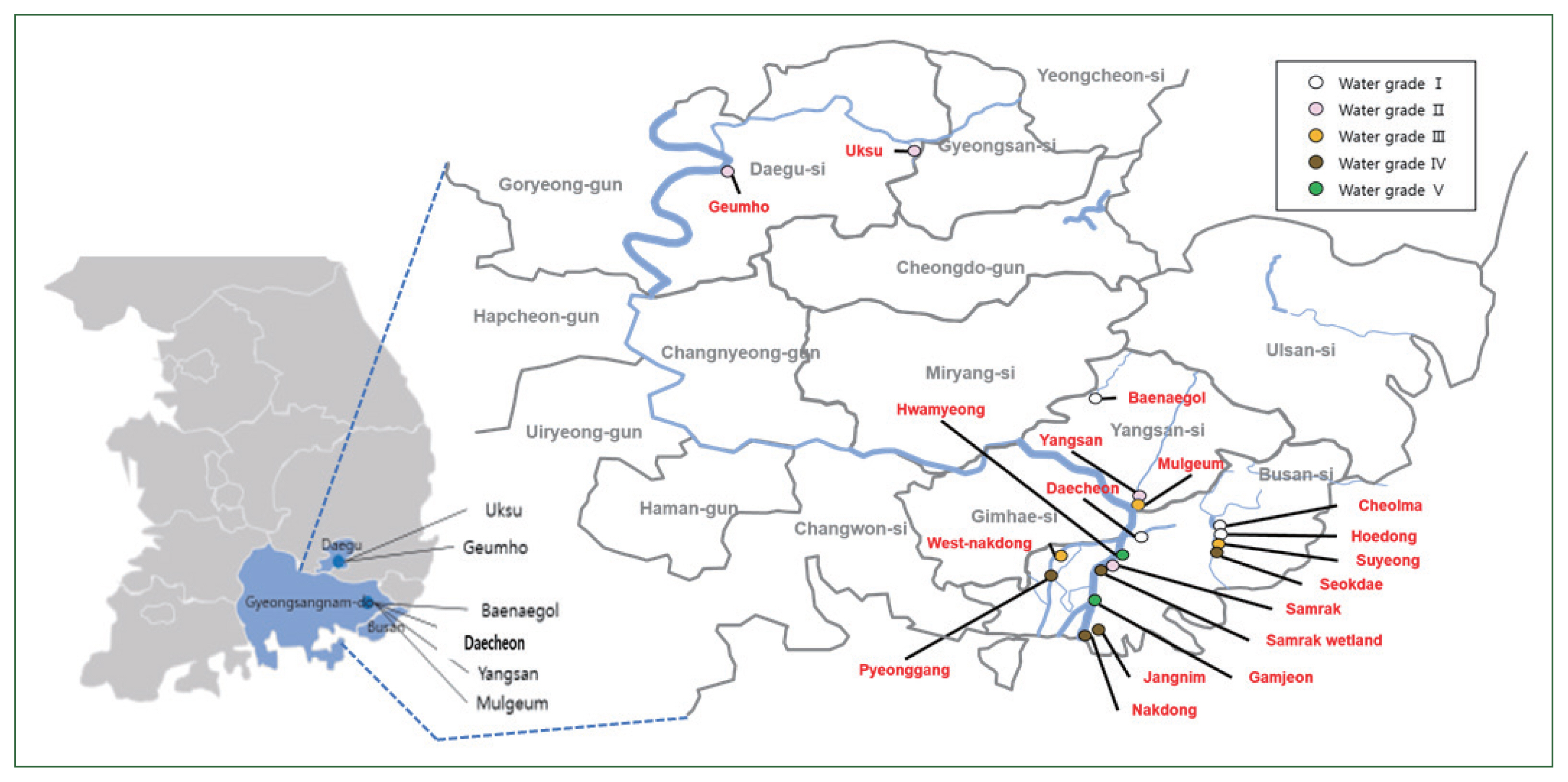
Fig. 2Time taken to microscopically detect the free-living amoebae (FLA) in monoxenic cultures. Comparisons of the earliest microscopic detection of FLA isolated from water samples of different quality. The line represents the trend line.
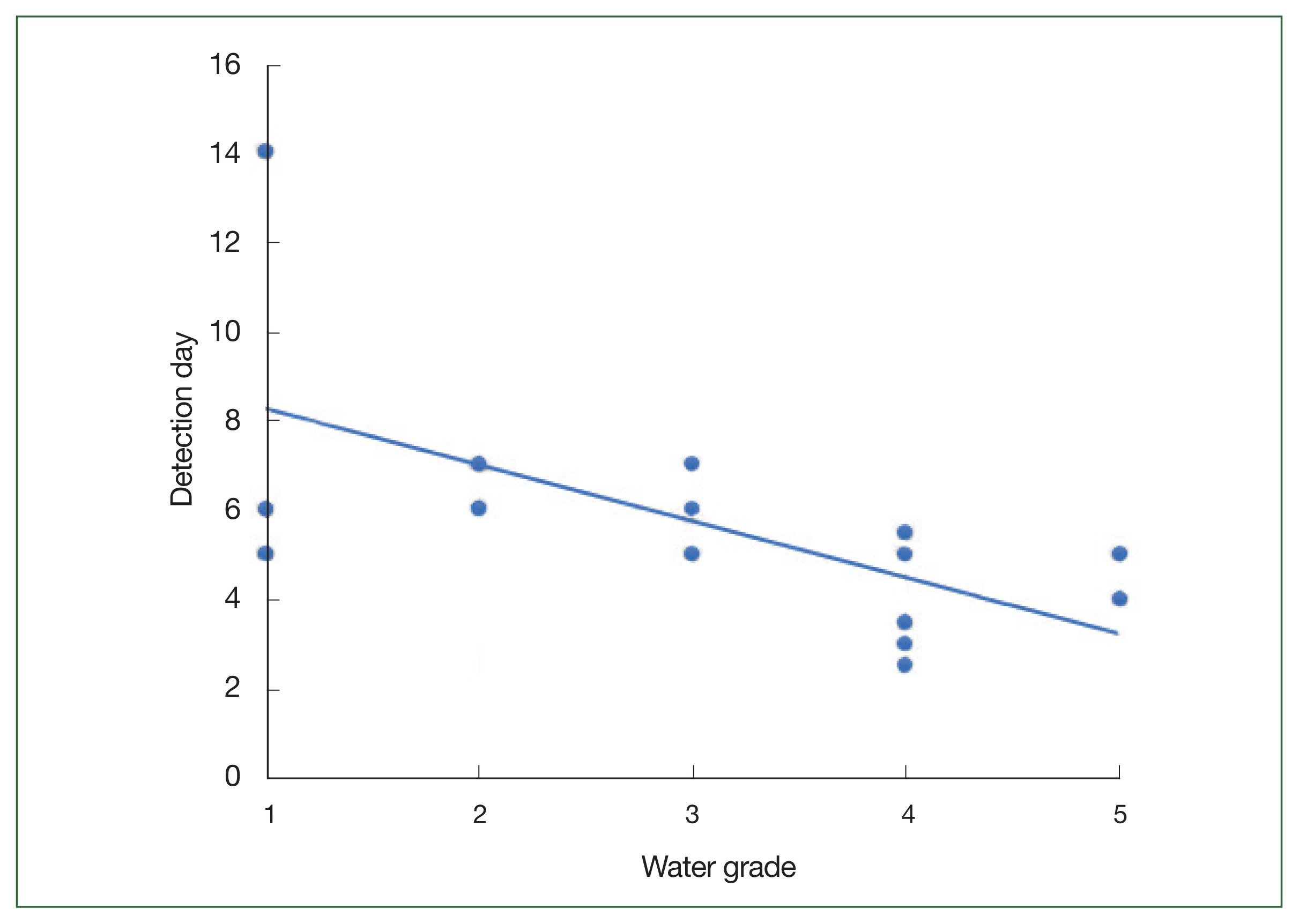
Fig. 3Genetic analysis of FLA isolates. A UPGMA dendrogram constructed based on the comparative 18S rDNA sequence analysis shows the relationships between FLA isolated from water samples in this study and the representative members of the Acanthamoeba, Vermamoeba, and Vannella genera. The amoebae isolated in this study and the quality of the water samples from which they were isolated are indicated in blue. The numbers in parentheses after each strain name indicate the GenBank accession numbers.
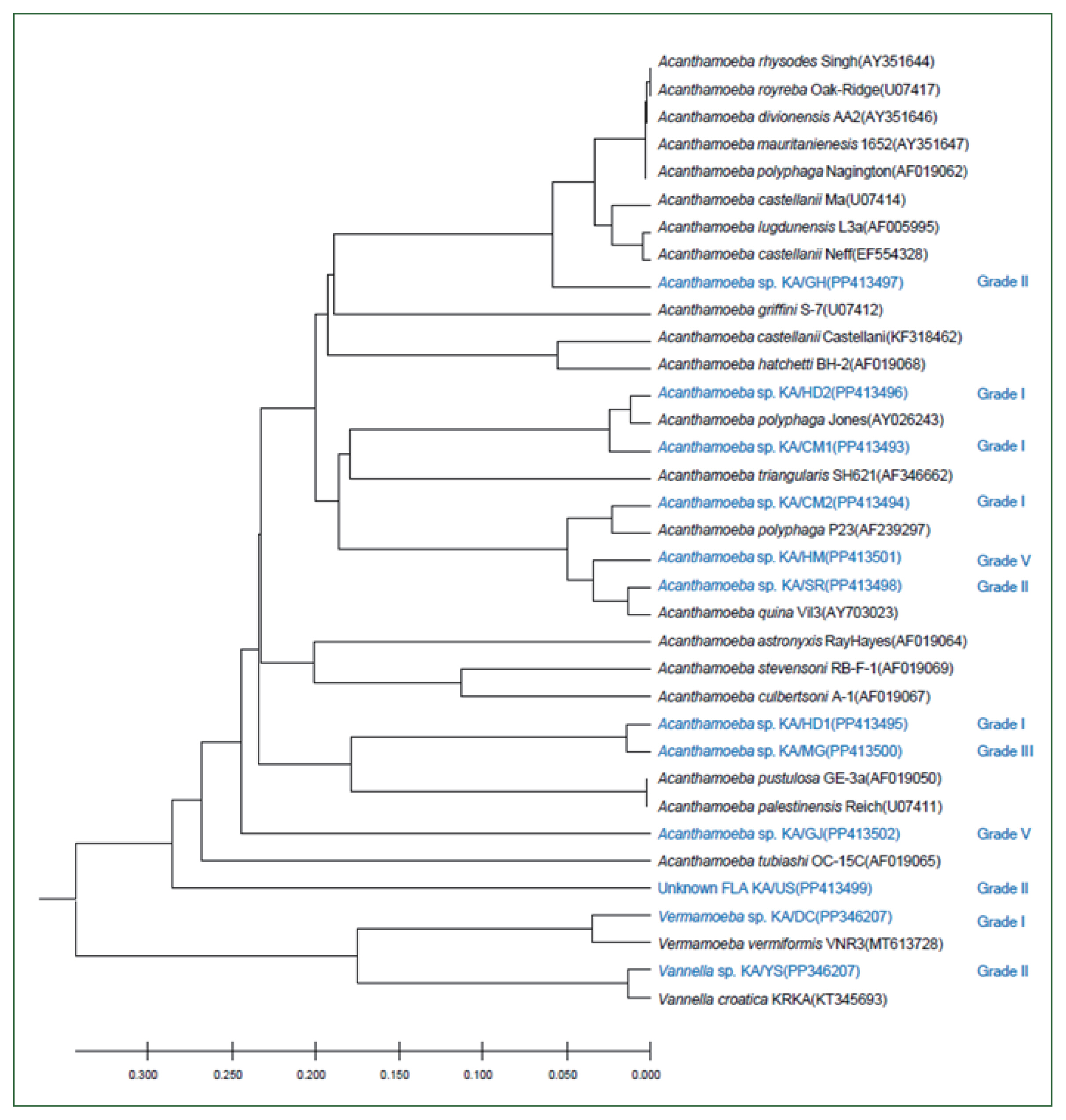
Fig. 4Genetic analysis of endosymbionts in the FLA isolates. A UPGMA dendrogram constructed based on the comparative 16S rDNA sequence analysis shows the relationships between endosymbionts in FLA isolated from water samples and previously isolated endosymbionts. The numbers in parentheses after each strain name indicate the GenBank accession numbers.
Fig. 5Morphological features of the endosymbiont in the FLA isolated in this study. The endosymbionts in the isolates KA/CM2 (A), KA/US (B), and KA/MG (C) are closely related to Candidatus Paracaedibacter acanthamoebae. The endosymbiont in the isolate KA/HD1 (D) is closely related to species in the Holosporaceae family; the endosymbiont in the isolate KA/SR (E) is closely related to species in the Sinorickettsia genus; and the endosymbiont in the isolate KA/HM (F) is closely related to species in the Holosporaceae family. No endosymbionts were detected in the KA/GJ isolate (G). Arrows indicate the endosymbionts. The red arrowheads indicate bacteria-like particles. N, nucleus; M, mitochondria.
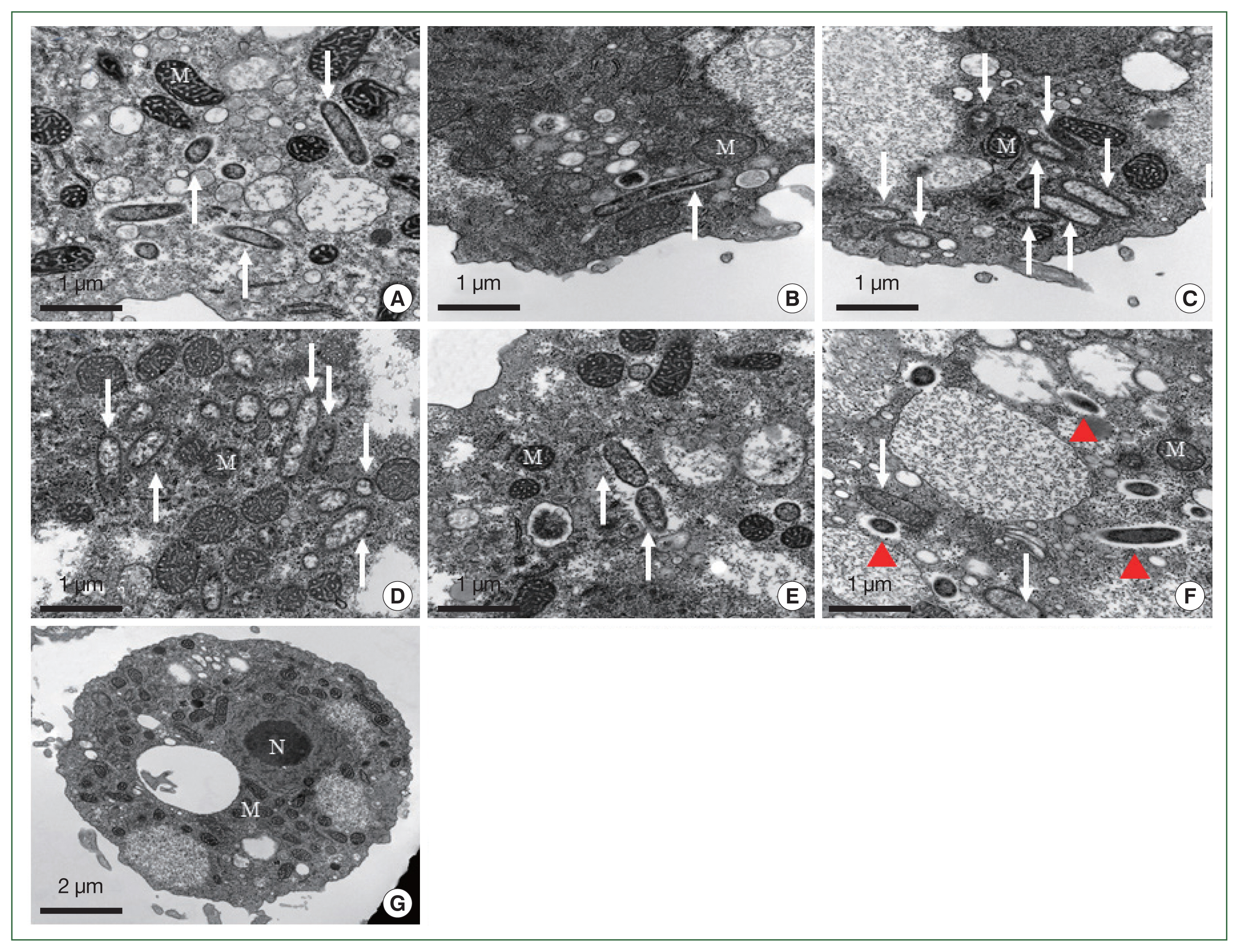
Table 1Information on the free-living amoeba isolated in this study
Table 1
|
Water quality |
Site |
BOD (mg/L) |
Total coliform
|
Fecal coliform
|
Isolate |
Cyst size (μm) |
Trophozoite size (μm) |
|
I |
Daecheon |
1.3 |
4,370 |
566 |
Va (KA/DC) |
7.8–8.3 |
7.3–10.0 |
|
Cheolma |
Ab (KA/CM1) |
14.0–18.5 |
20.0–26.0 |
|
A (KA/CM2) |
15.0–16.5 |
16.0–26.0 |
|
Hoedong |
A (KA/HD1) |
14.0–17.0 |
18.0–21.0 |
|
A (KA/HD2) |
14.0–17.5 |
15.0–22.0 |
|
Baenaegol |
Uc
|
- |
- |
|
|
II |
Geumho |
2.2 |
4,630 |
1,477 |
A (KA/GH) |
16.5–21.0 |
8.0–12.5 |
|
Samrak |
A (KA/SR) |
15.5–16.5 |
17.5–24.0 |
|
Yangsan |
Vcd (KA/YS) |
32.0–33.0 |
26.0–34.0 |
|
Uksu |
A (KA/US) |
11 |
13 |
|
|
III |
Mulgeum |
3.0 |
3,750 |
787 |
A (KA/MG) |
18.0–20.0 |
18.0–30.0 |
|
West-nakdong |
U |
- |
- |
|
Suyeong |
U |
- |
- |
|
|
IV |
Pyeonggang |
6.9 |
35,600 |
1,065 |
U |
- |
- |
|
Seokdae |
U |
- |
- |
|
Jangnim |
U |
- |
- |
|
Nakdong |
U |
- |
- |
|
Samrak-wetland |
U |
- |
- |
|
|
V |
Hwamyung |
13.2 |
17,000 |
4,200 |
A (KA/HM) |
11.0–14.0 |
16.0–21.0 |
|
Gamjeon |
A (KA/GJ) |
17 |
19.0–30.0 |
References
- 1. Prazeres M, Martínez-Colón M, Hallock P. Foraminifera as bioindicators of water quality: the FoRAM Index revisited. Environ Pollut 2020;257:113612. https://doi.org/10.1016/j.envpol.2019.113612
- 2. Cooper T, Gilmour J, Fabricius K. Bioindicators of changes in water quality on coral reefs: review and recommendations for monitoring programmes. Coral Reefs 2009;28(3):589-606. https://doi.org/10.1007/s00338-009-0512-x
- 3. Anseong-si K. River Water Quality Class Reference Table 2022 [Internet] Available from: https://www.anseong.go.kr/depart/contents.do?mId=0711030200#
- 4. Xuan Y, Shen Y, Ge Y, Yan G, Zheng S. Isolation and identification of Acanthamoeba strains from soil and tap water in Yanji, China. Environ Health Prev Med 2017;22(1):1-6. https://doi.org/10.1186/s12199-017-0655-2
- 5. Qin Y, Xie S, Smith HG, Swindles GT, Gu Y. Diversity, distribution and biogeography of testate amoebae in China: implications for ecological studies in Asia. Eur J Protistol 2011;47(1):1-9. https://doi.org/10.1016/j.ejop.2010.09.004
- 6. Roe HM, Patterson RT. Arcellacea (testate amoebae) as bio-indicators of road salt contamination in lakes. Microb Ecol 2014;68(2):299-313. https://doi.org/10.1007/s00248-014-0408-3
- 7. Nasser NA, Patterson RT, Roe HM, Galloway JM, Falck H, et al. Lacustrine arcellinina (testate amoebae) as bioindicators of arsenic contamination. Microb Ecol 2016;72(1):130-149. https://doi.org/10.1007/s00248-016-0752-6
- 8. Jeong HJ, Yu HS. The role of domestic tap water in Acanthamoeba contamination in contact lens storage cases in Korea. Parasites Hosts Dis 2005;43(2):47-50. https://doi.org/10.3347/kjp.2005.43.2.47
- 9. Lee WJ. New Korean records of two amoeboid protozoa (Protist); Vannella bursella and Pseudoparamoeba sp. J Species Res 2016;5(3):381-384.
- 10. Khwon WJ, Park JS. Morphology and phylogenetic analyses of three novel Naegleria isolated from freshwaters on Jeju Island, Korea, during the winter period. J Eukaryot Microbiol 2018;65(1):61-69. https://doi.org/10.1111/jeu.12434
- 11. Park JS. First record of potentially pathogenic amoeba Vermamoeba vermiformis (Lobosea: Gymnamoebia) isolated from a freshwater of Dokdo Island in the East Sea, Korea. Anim Syst Evol Divers 2016;32(1):1-8. https://doi.org/10.5635/ASED.2016.32.1.001
- 12. Cope JR, Landa J, Nethercut H, Collier SA, Glaser C, et al. The epidemiology and clinical features of Balamuthia mandrillaris disease in the United States, 1974–2016. Clin Infect Dis 2019;68(11):1815-1822. https://doi.org/10.1093/cid/ciy813
- 13. Lasjerdi Z, Niyyati M, Haghighi A, Zaeri F, Mojarad EN. First report of Vannellidae amoebae (Vannella spp.) isolated from biofilm source. Iran J Parasitol 2011;6(4):84.
- 14. Ettinger MR, Webb SR, Harris SA, McIninch SP, Garman CG, et al. Distribution of free-living amoebae in James River, Virginia, USA. Parasitol Res 2002;89(1):6-15. https://doi.org/10.1007/s00436-002-0707-3
- 15. Ma P, Visvesvara GS, Martinez AJ, Theodore FH, Daggett PM, et al. Naegleria and Acanthamoeba infections: review. Rev Infect Dis 1990;12(3):490-513. https://doi.org/10.1093/clinids/12.3.490
- 16. Grace E, Asbill S, Virga K. Naegleria fowleri: pathogenesis, diagnosis, and treatment options. Antimicrob Agents Chemother 2015;59(11):6677-6681. https://doi.org/10.1128/AAC.01293-15
- 17. Siddiqui R, Makhlouf Z, Khan NA. The increasing importance of Vermamoeba vermiformis. J Eukaryot Microbiol 2021;68(5):e12857. https://doi.org/10.1111/jeu.12857
- 18. Busan Metropolitan City. Waterquality [Internet];; [updated 2022 Aug 8]. Available from: https://www.busan.go.kr/depart/waterquality01
- 19. Water Environment Information System. Water quality. [Internet]; Available from: https://water.nier.go.kr/mobile/waterQuality?pMENU_NO=120
- 20. Jeong HJ, Lee SJ, Kim JH, Xuan YH, Lee KH, et al. Acanthamoeba: keratopathogenicity of isolates from domestic tap water in Korea. Exp Parasitol 2007;117(4):357-367. https://doi.org/10.1016/j.exppara.2007.05.003
- 21. Kim JY, Yi MH, Kim M, Yeom JS, Yoo HD, et al. Diagnosis of Balamuthia mandrillaris encephalitis by thymine-adenine cloning using universal eukaryotic primers. Ann Lab Med 2022;42(2):196-202. https://doi.org/10.3343/alm.2022.42.2.196
- 22. Smith VH, Tilman GD, Nekola JC. Eutrophication: impacts of excess nutrient inputs on freshwater, marine, and terrestrial ecosystems. Environ Pollut 1999;100(1–3):179-196. https://doi.org/10.1016/s0269-7491(99)00091-3
- 23. Smirnov AV, Bondarenko N, Glotova A, Nassonova E. Morphology and phylogeny of Vannella croatica n. sp. (Amoebozoa, Discosea, Vannellida). Eur J Protistol 2016;52:65-72. https://doi.org/10.1016/j.ejop.2015.11.002
- 24. Lee DI, Park SH, Baek JH, Yoon JW, Jin SI, et al. Identification of free-living amoebas in tap water of buildings with storage tanks in Korea. Parasites Hosts Dis 2020;58(2):191-194. https://doi.org/10.3347/kjp.2020.58.2.191
- 25. Lee DI, Park SH, Kang SA, Kim DH, Kim SH, et al. Free-Living Amoeba Vermamoeba vermiformis induces allergic airway inflammation. Parasites Hosts Dis 2022;60(4):229-239. https://doi.org/10.3347/kjp.2022.60.4.229
- 26. Castelli M, Lanzoni O, Giovannini M, Lebedeva N, Gammuto L, et al. ‘Candidatus Gromoviella agglomerans,, a novel intracellular Holosporaceae parasite of the ciliate Paramecium showing marked genome reduction. Environ Microbiol Rep 2022;14(1):34-49. https://doi.org/10.1111/1758-2229.13021
- 27. Xuan YH, Yu HS, Jeong HJ, Seol SY, Chung DI, et al. Molecular characterization of bacterial endosymbionts of Acanthamoeba isolates from infected corneas of Korean patients. Parasites Hosts Dis 2007;45(1):1-9. https://doi.org/10.3347/kjp.2007.45.1.1
- 28. Chidiac S, El Najjar P, Ouaini N, El Rayess Y, El Azzi D. A comprehensive review of water quality indices (WQIs): history, models, attempts and perspectives. Rev Environ Sci Biotechnol 2023;22(2):349-395. https://doi.org/10.1007/s11157-023-09650-7
- 29. Pathak D, Whitehead PG, Futter MN, Sinha R. Water quality assessment and catchment-scale nutrient flux modeling in the Ramganga River Basin in north India: an application of INCA model. Sci Total Environ 2018;631–632:201-215. https://doi.org/10.1016/j.scitotenv.2018.03.022
- 30. Fraga MS, Reis GB, da Silva DD, Guedes HAS, Elesbon AAA. Use of multivariate statistical methods to analyze the monitoring of surface water quality in the Doce River basin, Minas Gerais, Brazil. Environ Sci Pollut Res Int 2020;27(28):35303-35318. https://doi.org/10.1007/s11356-020-09783-0
 , Ji Won Seong2,†
, Ji Won Seong2,† , Jeong Hyun Kim2, Jun Young Lee2, Hyun Jae Cho2, Shin Ae Kang1, Mi Kyung Park1, Mi Jin Jeong1, Seo Yeong Choi1, Yu Jin Jeong1, Hak Sun Yu1,3,*
, Jeong Hyun Kim2, Jun Young Lee2, Hyun Jae Cho2, Shin Ae Kang1, Mi Kyung Park1, Mi Jin Jeong1, Seo Yeong Choi1, Yu Jin Jeong1, Hak Sun Yu1,3,*