Abstract
Caryophyllaeus brachycollis mainly parasitizes the intestines of globally distributed freshwater fishes, and infection causes significant economic losses to the aquaculture industry. However, data on the molecular epidemiology, population genetics, and systematics of C. brachycollis are scarce. In this study, we sequenced the complete mitogenome of C. brachycollis isolated from Beijing, China. This circular mitogenome comprised 14,273 bp, which was 231 bp shorter than that of C. brachycollis isolated from Wuhan, China. The mitogenome contained 12 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 2 noncoding regions. Bayesian inference revealed that C. brachycollis belonged to the family Caryophyllaeidae. The taxonomic status of C. brachycollis is controversial when based solely on morphological features. A comparative analysis of the mitogenome sequence obtained in this study revealed novel molecular markers for the accurate ascertainment of the phylogenetic position of this parasite.
-
Key words: Carp tapeworm, Caryophyllaeus brachycollis, mitochondrial genome, phylogenetic analyses
Introduction
Caryophyllideans, the earliest diverging group of non-segmented tapeworms, are described by morphological and biological features and typically have a monozoic (non-proglottized) body plan [
1–
3]. They are globally distributed and host-specific, mainly parasitizing in the intestinal tracts of Cypriniformes and Siluriformes fishes. Infection leads to weight loss, reduced fecundity, and high mortality rates, severely impacting the aquaculture industry [
4–
9]. The evolution of Eucestoda is unclear. The results of studies analyzing nuclear DNA genes (large and small subunits of ribosomal DNA) suggest that Spathebothriidea is the most divergent eucestode [
10,
11], whereas those analyzing morphological features and large fragments of mitochondrial DNA suggest that Caryophyllidea is the earliest divergent taxon [
3,
12]. Therefore, a better understanding of the relationship between orders Spathebothriidea and Caryophyllidea is warranted.
Although the order Caryophyllidea is regarded as a key group in Eucestoda research, the systematic classification and phylogenetic relationships of its members remain controversial. The families Balanotaeniidae, Capingentidae, Caryophyllaeidae, and Lytocestidae [
1,
2] are generally considered to belong to this order. Caryophyllaeidae, which frequently parasitize cyprinid fish in the Palearctic region, is the biggest group within Caryophyllidea, comprising 20 genera [
13]. However, a recent study suggested reclassification into 10 genera [
2], including
Caryophyllaeus (Gmelin, 1790). The number of species in this genus varies from the previously described 42 nominal taxa to the recently recognized 7 valid species [
14,
15], of which
Caryophyllaeus brachycollis is an important member.
C. brachycollis consists of 2 morphotypes based on morphological and molecular (large subunit of ribosomal DNA and
cox1) characteristics [
16,
17]. Although the mitogenome is a useful marker for tapeworm identification, differentiation, and systematic analyses [
18–
21], the complete mitogenome is only available for 1 morphotype, which was isolated from Wuhan, Hubei Province, China (CBCW). Because there is no data on the other morphotype, the molecular differentiation of
C. brachycollis in China is unclear.
Thus, our study focused on obtaining the complete mitogenome of the other C. brachycollis morphotype, which was isolated from Beijing, China (CBCB). The aims were as follows: (i) sequence and annotate the complete CBCB mitogenome, (ii) determine differences between CBCB and CBCW, (iii) infer the phylogenetic relationship among members of the order Caryophyllidea, and (iv) identify the systematic and taxonomic status of the genus Caryophyllaeus. These findings would provide more molecular data for use in further Caryophyllidea analyses.
Materials and Methods
Ethics statement
The experimental protocol was established according to the ethical guidelines of the Institutional Animal Care and Use Committee and approved by the Ethics Committee of Hunan Agricultural University (No. 202282).
Sample collection and DNA extraction
Samples were collected from the intestines of Cyprinus carpio from Beijing Zoo, China. They were washed in physiological saline, and those morphologically identified as caryophyllidean were fixed in 70% (v/v) ethanol and stored at −40°C until use.
Total genomic DNA was extracted using sodium dodecyl sulfate/proteinase K treatment, followed by spin-column purification (Wizard SV Genomic DNA Purification System; Promega, Madison, WI, USA). The purified DNA was used as the template in PCR assays with the primer pair JB3/JB4.5 targeting a fragment of
cox1 for amplification [
22].
A genomic DNA library (with 350 bp inserts) was prepared and sequenced by Novogene (Tianjin, China) using a HiSeq 2500 sequencing platform (Illumina, San Diego, CA, USA) to obtain 250 bp paired-end reads. Geneious v11.1.5 software was used to filter and clean the raw data and assemble the complete CBCB mitogenome using the amplified
cox1 sequences as initial references. The parameters were set as follows: minimum overlap identity, 99.0%; minimum overlap, 150 bp; and maximal gap size, 5 bp. The assembled mitogenome was further verified by long PCR experiments (
Supplementary Fig. S1) using the primers listed in
Supplementary Table S1.
The boundaries of protein-coding genes (PCGs) were identified using the NCBI tool ORF Finder (
https://www.ncbi.nlm.nih.gov/orffinder/). The initiation/termination codons for each PCG were identified following alignment with the CBCW mitogenome sequence using MAFFT v7.122 software. All PCG nucleotide sequences were translated to amino acid sequences using MEGA v11.0. Twenty-two transfer RNA (tRNA) genes were detected using ARWEN [
23] and tRNAscan-SE [
24]. Nucleotide composition and codon usage frequencies of the PCGs were analyzed using MEGA v11.0. The annotated CBCB mitogenome was visualized using the online server Proksee (
https://proksee.ca/). The CBCW sequence was used as the reference genome for comparisons with CBCB.
Phylogenetic analyses were performed using 6 mitogenome sequences of tapeworms within the order Caryophyllidea (
Table 1) and
Paruterina candelabraria (GenBank accession No. NC_039533) as the outgroup. Amino acid sequences were aligned using MAFFT v7.122 and concatenated into a single dataset. Ambiguous bases or gaps were excluded using Gblocks 0.91b software with the default parameters [
25]. Phylogenetic analyses were performed using Bayesian inference (BI) and maximum likelihood (ML) methods. BI analysis was run in MrBayes 3.1.1 using an approach similar to that of Ronquist and Huelsenbeck [
26]. ML analysis was performed using PhyML 3.1 software. “JTT+I+G+F” was selected as the best suitable model by ProtTest v3.4.2 based on the Akaike information criterion [
27] and was used for the BI and ML analyses. Phylograms were drawn using FigTree v.1.4.2 (
http://tree.bio.ed.ac.uk/software/figtree).
To determine the systematic relationship of the families in Caryophyllidea, BI analysis was performed using 20 cox1 sequences of the order Caryophyllidea and Gigantolina magna (GenBank accession No. JQ268545) as the outgroup.
Results
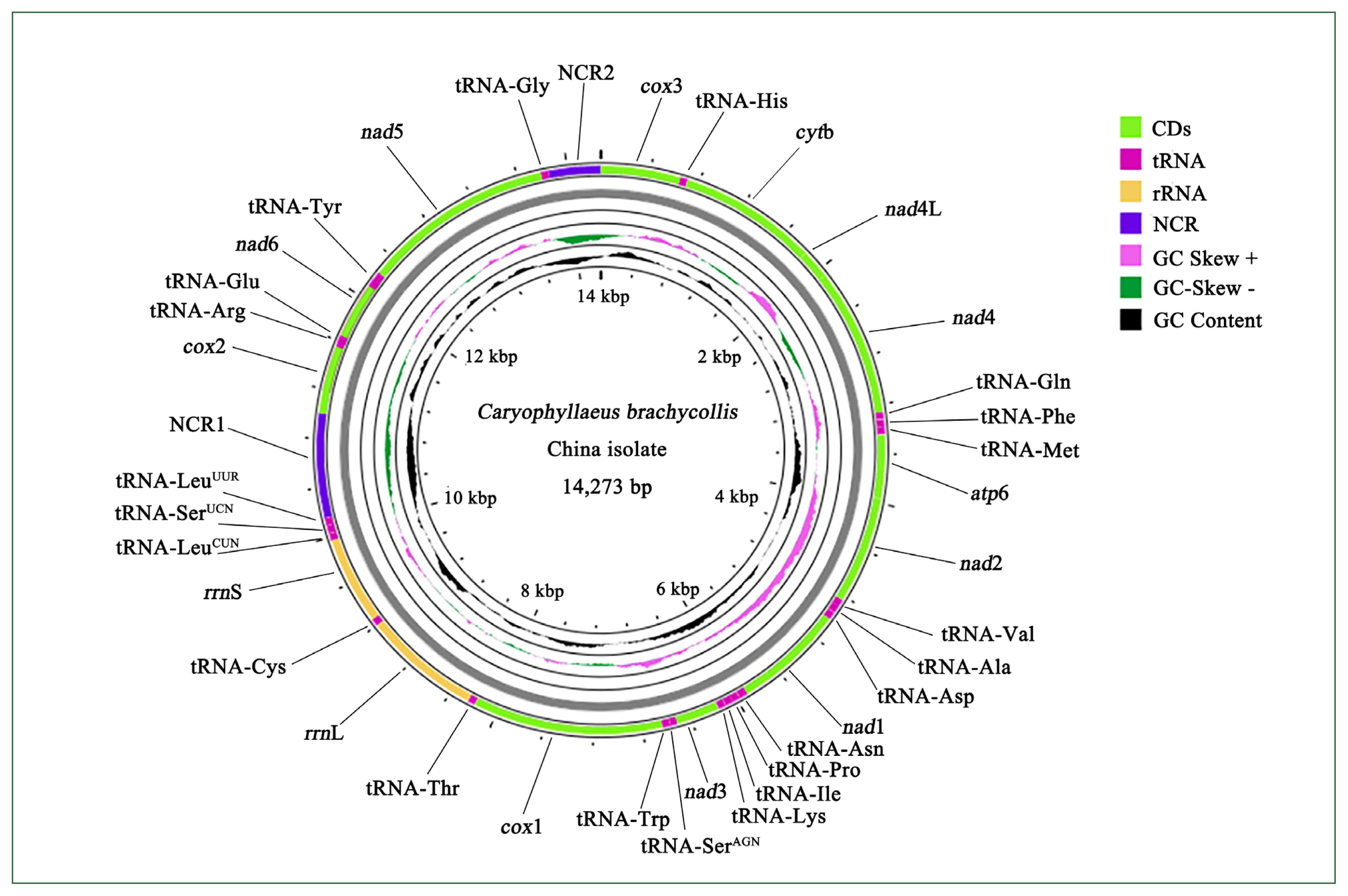
Genome composition and organization
Sequencing of the CBCB mitogenome generated 4.08 Gb of raw data, from which approximately 7,577,818×2 clean reads were obtained following filtering and used for further assembly. The complete circular CBCB mitogenome (GenBank accession No. OP359069) was 14,273 bp and comprised 36 genes, including 12 PCGs (excluding
atp8), 22 tRNA genes, 2 ribosomal RNA genes (
rrnS and
rrnL), and 2 noncoding regions (NCRs; control or AT-rich regions) (
Fig. 1;
Table 2). All 36 genes were transcribed in the forward direction. In
C. brachycollis mitochondrial DNA, the AT-skews were all negative (−0.481 to −0.125), and the GC-skews were all positive (0.167 to 0.465) (
Table 3), suggesting a bias toward T and G bases. The lengths of the 2 NCRs in CBCB were 845 and 423 bp, respectively, while those in CBCW were 641 and 871 bp, respectively (
Table 2). The large NCR (LNCR) was located between tRNA-Leu2 and
cox2, and the small NCR (SNCR) was located between tRNA-Gly and
cox3 (
Fig. 1). Alignment using BLAST revealed 100.0% nucleotide similarity with CBCW (GenBank accession No. NC_035430).
ATN was extensively used as the initiation codon, especially ATG. GTG is an uncommon start codon in tapeworms, but it was observed in
cytb,
nad1, and
nad4L in this study (
Table 2). Furthermore, TAA and TAG were frequently used as termination codons (TAG:
cytb,
atp6,
nad3,
nad4, and
nad4L; TAA:
nad6) (
Table 2). Incomplete stop codons T and TA were also prevalent in termination, with
cox1,
cox3, and
nad2 using T and
cox2,
nad1, and
nad5 using TA (
Table 2). The AT-skews of the 12 protein genes were all negative (−0.481 (
nad3) to −0.181 (
cox2)), and the GC-skews were all positive (0.167 (
cox2) to 0.465 (
nad4L)), suggesting a large T base bias in
nad3, a G base tendency in
nad4L, and a minor tendency of T+G bases in
cox2.
The 22 tRNA genes ranged from 54 to 68 bp (
Table 2).
rrnL and
rrnS were 951 and 712 bp, respectively, with an A+T content of 62.2% and 62.4%, respectively (
Table 3).
Comparisons between the CBCB and CBCW mitogenome sequences revealed that CBCB was approximately 231 bp shorter than CBCW. In both CBCB (
Fig. 1) and CBCW, the SNCR and LNCR were located in “tRNA-Gly and
cox3” and “tRNA-Leu2 and
cox2,” respectively. The size (
cox2 and
nad5) and the initiation/termination (
cox2) codons of PCGs in CBCB and CBCW differed (
Table 2).
cox2 was 539 bp and 551 bp in CBCW and CBCB, respectively, while
nad5 was 1,547 and 1,550 bp, respectively. The initiation/termination codons of
cox2 were ATG/TAG and ATG/TA in CBCW and CBCB, respectively.
Comparison of the nucleotide sequences of PCGs between CBCB and CBCW revealed differences of 0% (
nad4L) to 2.2% (
cox3) (
Table 4). Differences in the amino acid sequences of CBCB and CBCW ranged from 0% (
nad4L) to 2.7% (
cox2) (
Table 4).
nad4L typically shows interspecies variability. However, our study revealed that it was highly conserved in
C. brachycollis. These findings suggested that
cox2 might be a better molecular marker for identifying cryptic species within
C. brachycollis.
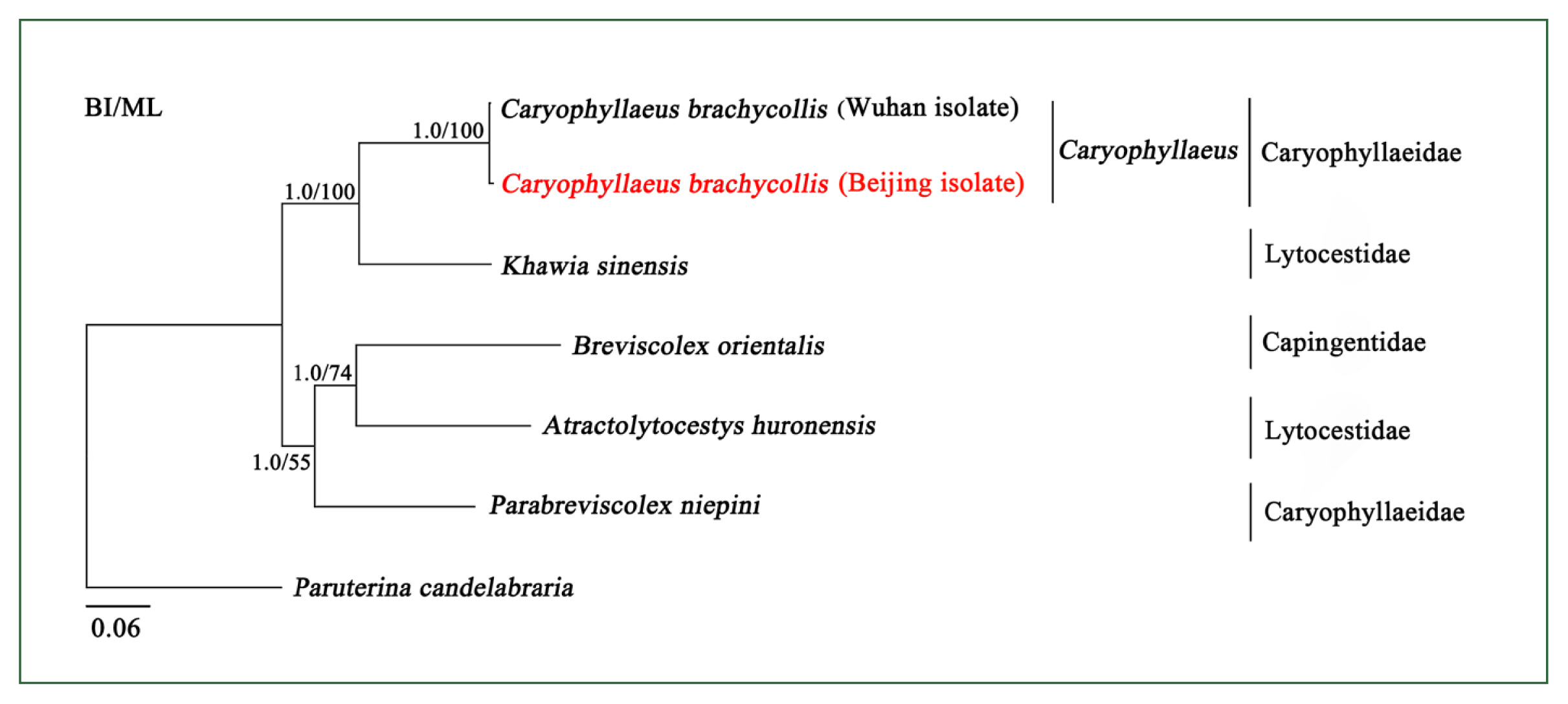
The phylogeny of Caryophyllidea is currently unclear due to a lack of complete mitogenome sequences. To better clarify the phylogenetic relationships among the members of this order, 6 mitogenome sequences were phylogenetically analyzed using BI and ML methods. The BI and ML tree topologies were identical (
Fig. 2). There were 2 main clades: 1 clade contained Caryophyllaeidae and Lytocestidae, and the other clade contained Capingentidae, Caryophyllaeidae, and Lytocestidae, indicating that families Caryophyllaeidae and Lytocestidae were paraphyletic. The mitochondrial DNA sequences of CBCB and CBCW grouped with short topological distance (BPP =1, BF=100) (
Fig. 2), indicating that they were closely related. Furthermore, phylogenetic analyses of
cox1 also showed that Caryophyllaeidae and Lytocestidae were paraphyletic and revealed a close evolutionary distance between CBCB and CBCW (
Supplementary Fig. S2).
Discussion
The complete mitogenome of CBCB was 14,273 bp and encoded 36 genes. The A+T base bias (73.2%) was significantly higher than the G+C base bias (26.8%). The AT-skew ([A-T]/[A+T]) and GC-skew ([G-C]/[G+C]) were useful methods to estimate compositional asymmetry [
28].
Regarding the initiation codons, ATG was the most prevalent in 8 of the 12 PCGs (
atp6,
cox1,
cox2,
nad2,
nad3,
nad4,
nad5, and
nad6), and ATT was used in
cox3. TAA and TAG have been reported as common termination codons [
29,
30] and were identified as frequent termination codons in our study. There were 22 tRNA genes in the CBCB mitogenome. Similar to tapeworms
Breviscolex orientalis and
Parabreviscolex niepini [
31,
32], most tRNA genes in CBCB presented as a typical ‘cloverleaf’ structure, excluding tRNA-Ser
AGN and tRNA-Arg, which lacked DHU arms. The
rrnL was located between tRNA-Thr and tRNA-Cys, and
rrnS between tRNA-Cys and tRNA-Leu1, similar to the arrangement in
B. orientalis and
Atractolytocestus huronensis [
32].
Although the mitogenome sequence of CBCW was longer than that of CBCB, the arrangements of the 36 genes were the same and consistent with the order observed in most caryophyllideans [
31,
32]. Considering that the differences in mitogenome sequences between closely related nematode species are 10.0%–20.0% [
33,
34], and species-level variations in mammals are typically 2.0%–10.0% [
35], the level of differences between CBCB and CBCW indicated that they were identical.
Comparison between the CBCB and CBCW mitogenomes revealed that cox2 is a reliable molecular marker for cryptic species identification. Furthermore, nad4L was highly conserved in C. brachycollis. This comparative mitogenomic analysis elucidated evolutionary and phylogenetic relationships among related species, enhancing the accuracy of taxonomic classification.
To summarize, our study determined and characterized the complete mitogenome sequence (14,273 bp) of CBCB, revealing that it was shorter than that of CBCW. The CBCB mitogenome provided evidence supporting a close phylogenetic relationship between C. brachycollis and other members of the Caryophyllidae family. Because only a limited number of specimens from a single geographic location were analyzed, future studies should include a broader sample to further assess intraspecific variations.
Notes
-
Author contributions
Conceptualization: Liu YL, Fu YT
Data curation: Liu YL, Zhang Y, Fu YT
Formal analysis: Zhang Y
Investigation: Liu YL
Methodology: Deng YP
Resources: Wang HM, Deng YP
Software: Wang HM
Supervision: Fu YT, Liu GH
Validation: Zhang Y, Fu YT, Liu GH
Writing – original draft: Liu YL, Zhang Y, Wang HM
Writing – review & editing: Deng YP
-
Conflict of interest
The authors declare no conflict of interest related to this study.
-
Acknowledgments
This study was supported in part, by the Training Programme for Excellent Young Innovators of Changsha (grant No. KQ2106044). The raw data have been deposited in GenBank (accession No. OP359069 and SRR34793993).
Supplementary Information
Fig. 1Mitogenome organization of Caryophyllaeus brachycollis isolate from China. Gene scaling is approximate. CD, coding region; NCR1, long noncoding region; NCR2, short noncoding region; tRNA, transfer RNA; rRNA, ribosomal RNA.
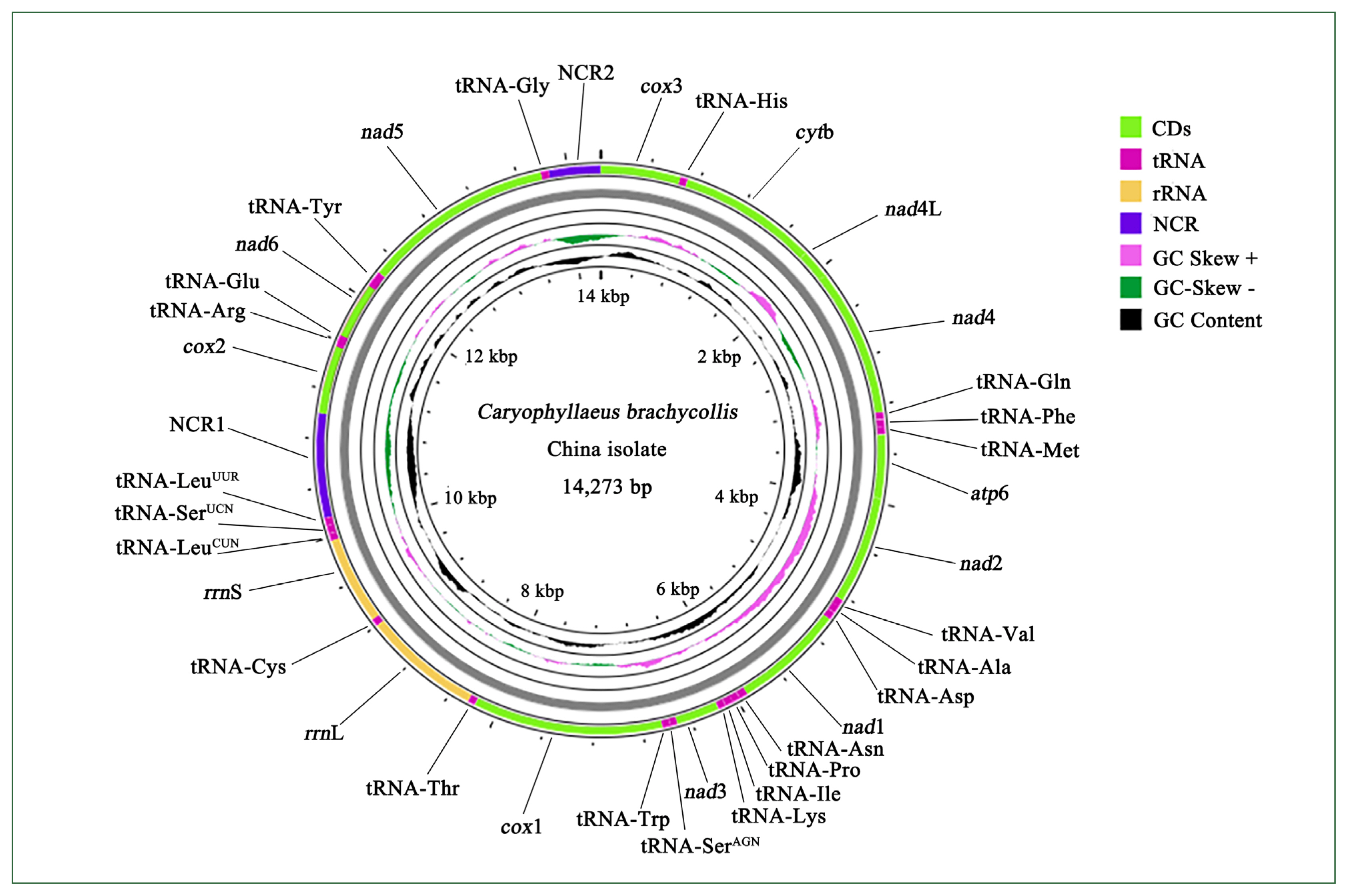
Fig. 2Inferred phylogenetic relationships among species in the order Caryophyllidea. The concatenated amino acid sequences of 12 mitochondrial protein-coding genes were analyzed utilizing maximum likelihood (ML) and Bayesian analysis (BI) methods, with Paruterina candelabraria as the outgroup. The Caryophyllaeus brachycollis form was isolated from carp tapeworm in Beijing in this study.
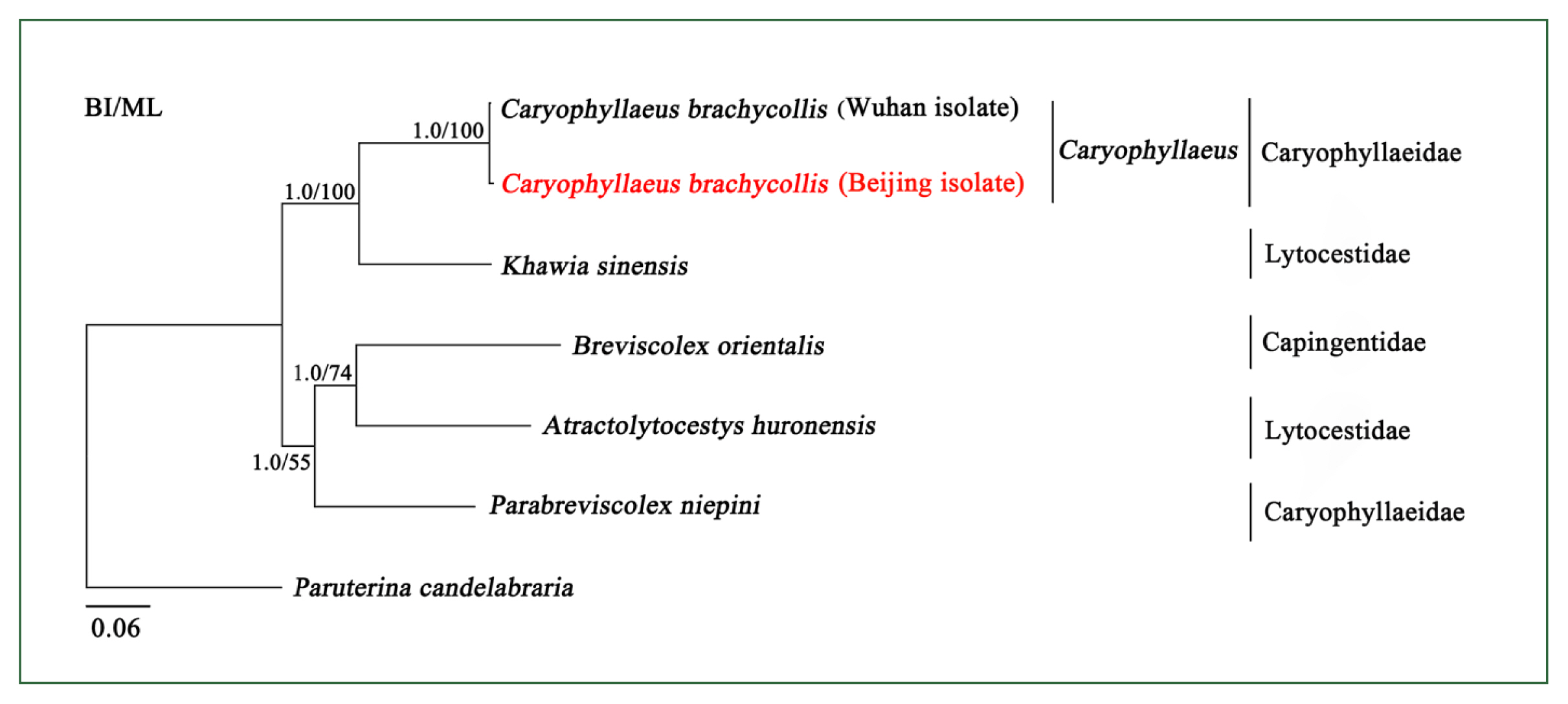
Table 1Complete mitogenome sequences of the order Caryophyllidea used for phylogenetic analysis
Table 1
|
Family/order |
Species |
Size (bp) |
GenBank accession No. |
|
Lytocestidae |
Khawia sinensis
|
13,759 |
NC034800 |
|
Atractolytocestys huronensis
|
15,130 |
NC035635 |
|
|
Caryophyllaeidae |
Caryophyllaeus brachycollis
|
14,504 |
NC035430 |
|
Parabreviscolex niepini
|
15,034 |
NC040117 |
|
|
Capingentidae |
Breviscolex orientalis
|
14,011 |
NC035634 |
|
|
Cyclophyllidea |
Paruterina candelabraria
|
13,634 |
NC039533 |
Table 2Comparative organization of the complete mitogenome of Caryophyllaeus brachycollis isolates from Beijing and Wuhan, China
Table 2
|
Gene/region |
Positions |
Size (bp) |
No. of aa |
Ini/Ter codons |
Anticodons |
|
|
|
|
|
|
CBCB |
CBCW |
CBCB |
CBCW |
CBCB |
CBCW |
CBCB |
CBCW |
CBCB/CBCW |
|
cox3
|
1–643 |
1–643 |
643 |
643 |
214 |
214 |
ATT/T |
ATT/T |
|
|
|
tRNA-His (H) |
644–705 |
644–705 |
62 |
62 |
|
|
|
|
GTG |
|
|
cytb
|
707–1825 |
707–1825 |
1,119 |
1,119 |
372 |
372 |
GTG/TAG |
GTG/TAG |
|
|
|
nad4L
|
1828–2067 |
1828–2067 |
240 |
240 |
79 |
79 |
GTG/TAG |
GTG/TAG |
|
|
|
nad4
|
2028–3260 |
2028–3260 |
1,233 |
1,233 |
410 |
410 |
ATG/TAG |
ATG/TAG |
|
|
|
tRNA-Gln (Q) |
3264–3318 |
3261–3322 |
55 |
63 |
|
|
|
|
TTG |
|
|
tRNA-Phe (F) |
3323–3377 |
3320–3382 |
55 |
63 |
|
|
|
|
GAA |
|
|
tRNA-Met (M) |
3378–3440 |
3378–3440 |
63 |
63 |
|
|
|
|
CAT |
|
|
atp6
|
3444–3959 |
3444–3959 |
516 |
516 |
171 |
171 |
ATG/TAG |
ATG/TAG |
|
|
|
nad2
|
3960–4830 |
3960–4830 |
871 |
871 |
290 |
290 |
ATG/T |
ATG/T |
|
|
|
tRNA-Val (V) |
4831–4890 |
4831–4890 |
60 |
60 |
|
|
|
|
TAC |
|
|
tRNA-Ala (A) |
4889–4949 |
4888–4949 |
61 |
62 |
|
|
|
|
TGC |
|
|
tRNA-Asp (D) |
4954–5014 |
4954–5014 |
61 |
61 |
|
|
|
|
GTC |
|
|
nad1
|
5015–5907 |
5015–5908 |
893 |
894 |
297 |
297 |
GTG/TA |
GTG/TAG |
|
|
|
tRNA-Asn (N) |
5908–5975 |
5908–5975 |
68 |
68 |
|
|
|
|
GTT |
|
|
tRNA-Pro (P) |
5980–6040 |
5980–6040 |
61 |
61 |
|
|
|
|
TGG |
|
|
tRNA-Ile (I) |
6040–6103 |
6041–6102 |
64 |
62 |
|
|
|
|
GAT |
|
|
tRNA-Lys (K) |
6106–6165 |
6106–6165 |
60 |
60 |
|
|
|
|
GTT |
|
|
nad3
|
6170–6517 |
6170–6517 |
348 |
348 |
115 |
115 |
ATG/TAG |
ATG/TAG |
|
|
|
tRNA-Ser1 (S1) |
6520–6574 |
6518–6575 |
55 |
58 |
|
|
|
|
GCT |
|
|
tRNA-Trp (W) |
6575–6638 |
6575–6638 |
64 |
64 |
|
|
|
|
TCA |
|
|
cox1
|
6642–8178 |
6642–8201 |
1,537 |
1,537 |
512 |
512 |
ATG/T |
ATG/T |
|
|
|
tRNA-Thr (T) |
8183–8242 |
8183–8242 |
60 |
60 |
|
|
|
|
TGT |
|
|
rrnL
|
8243–9193 |
8238–9191 |
951 |
954 |
|
|
|
|
|
|
|
tRNA-Cys |
9194–9254 |
9192–9252 |
61 |
61 |
|
|
|
|
GCA |
|
|
rrnS
|
9255–9966 |
9253–9965 |
712 |
713 |
|
|
|
|
|
|
|
tRNA-Leu1 (L1) |
9967–10028 |
9966–10027 |
62 |
62 |
|
|
|
|
TAG |
|
|
tRNA-Ser2 (S2) |
10030–10093 |
10028–10093 |
64 |
66 |
|
|
|
|
TGA |
|
|
tRNA-Leu2 (L2) |
10095–10157 |
10094–10156 |
63 |
63 |
|
|
|
|
TAA |
|
|
NCR1 |
10158–11002 |
10157–10797 |
845 |
641 |
|
|
|
|
|
|
|
cox2
|
11003–11553 |
10798–11336 |
551 |
539 |
183 |
179 |
ATG/TA |
ATG/TAG |
|
|
|
tRNA-Glu (E) |
11587–11646 |
11370–11429 |
60 |
60 |
|
|
|
|
TTC |
|
|
nad6
|
11647–12105 |
11430–11888 |
459 |
459 |
152 |
152 |
ATG/TAA |
ATG/TAA |
|
|
|
tRNA-Tyr (Y) |
12113–12174 |
11896–11957 |
62 |
62 |
|
|
|
|
GTA |
|
|
tRNA-Arg (R) |
11183–12236 |
11966–12021 |
54 |
56 |
|
|
|
|
TCG |
|
|
nad5
|
12237–13786 |
12023–13570 |
1,550 |
1,547 |
516 |
515 |
ATG/TA |
ATG/TA |
|
|
|
tRNA-Gly (G) |
13787–13850 |
13570–13633 |
64 |
64 |
|
|
|
|
TCC |
|
|
NCR2 |
13851–14273 |
13634–14504 |
423 |
871 |
|
|
|
|
|
Table 3Nucleotide composition and skews of the Caryophyllaeus brachycollis mitogenome
Table 3
|
Gene |
Nucleotide frequency |
A+T (%) |
AT-skew |
GC-skew |
|
A (%) |
G (%) |
T (%) |
C (%) |
|
atp6
|
16.7 |
25.0 |
42.2 |
16.1 |
58.9 |
−0.434 |
0.297 |
|
cox1
|
21.7 |
23.0 |
40.3 |
15.0 |
62.0 |
−0.230 |
0.212 |
|
cox2
|
25.4 |
22.1 |
36.7 |
15.8 |
62.1 |
−0.181 |
0.167 |
|
cox3
|
18.7 |
24.9 |
40.4 |
16.0 |
59.1 |
−0.368 |
0.217 |
|
cytb
|
22.0 |
23.2 |
39.9 |
14.9 |
61.9 |
−0.289 |
0.218 |
|
nad1
|
19.6 |
25.9 |
41.8 |
12.8 |
61.4 |
−0.361 |
0.339 |
|
nad2
|
16.6 |
26.5 |
45.8 |
11.0 |
62.4 |
−0.467 |
0.411 |
|
nad3
|
16.1 |
25.3 |
46.0 |
12.6 |
62.1 |
−0.481 |
0.333 |
|
nad4
|
20.8 |
22.4 |
41.7 |
15.1 |
62.5 |
−0.335 |
0.195 |
|
nad4L
|
19.6 |
26.2 |
44.6 |
9.6 |
64.2 |
−0.390 |
0.465 |
|
nad5
|
20.1 |
23.4 |
42.5 |
13.9 |
62.6 |
−0.357 |
0.254 |
|
nad6
|
20.0 |
22.9 |
42.3 |
14.8 |
62.3 |
−0.357 |
0.214 |
|
rrnS
|
27.0 |
23.9 |
35.4 |
13.8 |
62.4 |
−0.135 |
0.269 |
|
rrnL
|
27.2 |
22.7 |
35.0 |
15.0 |
62.2 |
−0.125 |
0.203 |
|
22 tRNAs |
25.9 |
24.8 |
34.3 |
15.0 |
60.2 |
−0.139 |
0.246 |
|
Total |
23.7 |
22.6 |
39.5 |
14.2 |
63.2 |
−0.250 |
0.227 |
Table 4Nucleotide and amino acid sequence differences between the isolates from Beijing, China and Wuhan, China mitogenomes
Table 4
|
Gene/region |
NT sequence length |
NT difference (%) |
No. of AA |
AA difference (%) |
|
|
|
|
|
CBCB |
CBCW |
CBCB/CBCW |
CBCB |
CBCW |
CBCB/CBCW |
|
atp6
|
516 |
516 |
1.7 |
171 |
171 |
1.2 |
|
|
cox1
|
1,537 |
1,537 |
0.8 |
512 |
512 |
1.0 |
|
|
cox2
|
551 |
539 |
1.5 |
183 |
179 |
2.7 |
|
|
cox3
|
643 |
643 |
2.2 |
214 |
214 |
1.4 |
|
|
cytb
|
1,119 |
1,119 |
1.3 |
372 |
372 |
0.3 |
|
|
nad1
|
893 |
894 |
0.6 |
297 |
297 |
0.7 |
|
|
nad2
|
871 |
871 |
0.6 |
290 |
290 |
0.3 |
|
|
nad3
|
348 |
348 |
0.3 |
115 |
115 |
0.9 |
|
|
nad4
|
1,233 |
1,233 |
1.4 |
397 |
410 |
0.7 |
|
|
nad4L
|
240 |
240 |
0.0 |
79 |
79 |
0.0 |
|
|
nad5
|
1,550 |
1,547 |
1.0 |
516 |
515 |
0.8 |
|
|
nad6
|
459 |
459 |
1.1 |
152 |
152 |
1.3 |
|
|
rrnS
|
712 |
713 |
1.7 |
- |
- |
- |
|
|
rrnL
|
951 |
954 |
0.8 |
- |
- |
- |
|
|
All 22tRNAs |
1,339 |
1,361 |
2.1 |
- |
- |
- |
References
- 1. Oros M, Hanzelová V, Scholz T, Mackiewicz JS. Phylogenetic relationships of the monozoic tapeworms (Eucestoda: Caryophyllidea) inferred from morphological characters. Syst Parasitol 2008;70(1):1-14. https://doi.org/10.1007/s11230-008-9133-y
- 2. Scholz T, Waeschenbach A, Oros M, Brabec J, Littlewood DTJ. Phylogenetic reconstruction of early diverging tapeworms (Cestoda: Caryophyllidea) reveals ancient radiations in vertebrate hosts and biogeographic regions. Int J Parasitol 2021;51(4):263-277. https://doi.org/10.1016/j.ijpara.2020.09.009
- 3. Waeschenbach A, Webster BL, Littlewood DTJ. Adding resolution to ordinal level relationships of tapeworms (Platyhelminthes: Cestoda) with large fragments of mtDNA. Mol Phylogenet Evol 2012;63(3):834-847. https://doi.org/10.1016/j.ympev.2012.02.020
- 4. Bruňanská M. Spermatological characters of the caryophyllidean cestode Khawia sinensis Hsü, 1935, a carp parasite. Parasitol Res 2009;105(6):1603-1610. https://doi.org/10.1007/s00436-009-1599-2
- 5. Králová-Hromadová I, Štefka J, Bazsalovicsová E, Bokorová S, Oros M. The tapeworm Atractolytocestus tenuicollis (Cestoda: Caryophyllidea): a sister species or ancestor of an invasive A. huronensis? Parasitol Res 2013;112(10):3379-3388. https://doi.org/10.1007/s00436-013-3516-y
- 6. Feng Y, Feng HL, Fang YH, Su YB. Characterization of the complete mitochondrial genome of Khawia sinensis belongs among platyhelminths, cestodes. Exp Parasitol 2017;177:35-39. https://doi.org/10.1016/j.exppara.2017.04.005
- 7. Oros M, Hanzelová V, Scholz T. Tapeworm Khawia sinensis: review of the introduction and subsequent decline of a pathogen of carp, Cyprinus carpio. Vet Parasitol 2009;164(2–4):217-222. https://doi.org/10.1016/j.vetpar.2009.05.010
- 8. Chisholm LA, Morgan JA, Adlard RD, Whittington ID. Phylogenetic analysis of the Monocotylidae (Monogenea) inferred from 28S rDNA sequences. Int J Parasitol 2001;31(11):1253-1263. https://doi.org/10.1016/S0020-7519(01)00223-5
- 9. Edema CU, Okaka CE, Oboh IP, Okogub BO. A preliminary study of parasitic infections of some fishes from Okhuo River, Benin City, Nigeria. Int J Biomed Health Sci 2008;4(3):107-112.
- 10. Olson PD, Caira JN. Evolution of the major lineages of tapeworms (Platyhelminthes: Cestoidea) inferred from 18S ribosomal DNA and elongation factor-1alpha. J Parasitol 1999;85(6):1134-1159. https://doi.org/10.2307/3285679
- 11. Waeschenbach A, Webster BL, Bray RA, Littlewood DTJ. Added resolution among ordinal level relationships of tapeworms (Platyhelminthes: Cestoda) with complete small and large subunit nuclear ribosomal RNA genes. Mol Phylogenet Evol 2007;45(1):311-325. https://doi.org/10.1016/j.ympev.2007.03.019
- 12. Hoberg EP, Mariaux J, Justine JL, Brooks DR, Weekes PJ. Phylogeny of the orders of the Eucestoda (Cercomeromorphae) based on comparative morphology: historical perspectives and a new working hypothesis. J Parasitol 1997;83(6):1128-1147. https://doi.org/10.2307/3284374
- 13. Protasova EP, Kuperman BI, Roitman VA, Poddubnaya LG. Caryophyllid Tapeworms of the Fauna of USSR. Nauka, Moscow. USSR; 1990, p 237.
- 14. Bazsalovicsová E, Králová-Hromadová I, Juhásová L, Mikulíček P, Oravcová A, et al. Comparative analysis of monozoic fish tapeworms Caryophyllaeus laticeps (Pallas, 1781) and recently described Caryophyllaeus chondrostomi Barčák, Oros, Hanzelová, Scholz, 2017, using microsatellite markers. Parasitol Res 2020;119(12):3995-4004. https://doi.org/10.1007/s00436-020-06898-8
- 15. Barcak D, Oros M, Hanzelova V, Scholz T. A synoptic review of Caryophyllaeus Gmelin, 1790 (Cestoda: Caryophyllidea), parasites of cyprinid fishes. Folia Parasitol(Praha); 2017. 64(2017):027 https://doi.org/10.14411/fp.2017.027
- 16. Barčák D, Oros M, Hanzelová V, Scholz T. Phenotypic plasticity in Caryophyllaeus brachycollis Janiszewska, 1953 (Cestoda: Caryophyllidea): does fish host play a role? Syst Parasitol 2014;88(2):153-166. https://doi.org/10.1007/s11230-014-9495-2
- 17. Bazsalovicsová E, Králová-Hromadová I, Brabec J, Hanzelová V, Oros M, et al. Conflict between morphology and molecular data: a case of the genus Caryophyllaeus (Cestoda: Caryophyllidea), monozoic tapeworms of cyprinid fishes. Folia Parasitol (Praha) 2014;61(4):347-354. https://doi.org/10.14411/fp.2014.035
- 18. Liu GH, Lin RQ, Li MW, Liu W, Liu Y, et al. The complete mitochondrial genomes of three cestode species of Taenia infecting animals and humans. Mol Biol Rep 2011;38(4):2249-2256. https://doi.org/10.1007/s11033-010-0355-0
- 19. Jeon HK, Eom KS. Molecular approaches to Taenia asiatica. Korean J Parasitol 2013;51(1):1-8. https://doi.org/10.3347/kjp.2013.51.1.1
- 20. Zhao GH, Wang HB, Jia YQ, Zhao W, Hu XF, et al. The complete mitochondrial genome of Pseudanoplocephala crawfordi and a comparison with closely related cestode species. J Helminthol 2016;90(5):588-595. https://doi.org/10.1017/S0022149X15000802
- 21. Li WX, Fu PP, Zhang D, Boyce K, Xi BW, et al. Comparative mitogenomics supports synonymy of the genera Ligula and Digramma (Cestoda: Diphyllobothriidae). Parasit Vectors 2018;11(1):324. https://doi.org/10.1186/s13071-018-2910-9
- 22. Simsek S, Cevik A. First detection and molecular characterization of Echinococcus equinus in a mule in Turkey. Acta Parasitol 2014;59(4):773-777. https://doi.org/10.2478/s11686-014-0308-1
- 23. Laslett D, Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008;24(2):172-175. https://doi.org/10.1093/bioinformatics/btm573
- 24. Chan PP, Lowe TM. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol 2019;1962:1-14. https://doi.org/10.1007/978-1-4939-9173-0_1
- 25. Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 2007;56(4):564-577. https://doi.org/10.1080/10635150701472164
- 26. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003;19(12):1572-1574. https://doi.org/10.1093/bioinformatics/btg180
- 27. Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 2011;27(8):1164-1165. https://doi.org/10.1093/bioinformatics/btr088
- 28. Reyes A, Gissi C, Pesole G, Saccone C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol 1998;15(8):957-966. https://doi.org/10.1093/oxfordjournals.molbev.a026011
- 29. Nakao M, Sako Y, Ito A. The mitochondrial genome of the tapeworm Taenia solium: a finding of the abbreviated stop codon U. J Parasitol 2003;89(3):633-635. https://doi.org/10.1645/0022-3395(2003)089[0633:TMGOTT]2.0.CO;2
- 30. Jeon HK, Kim KH, Eom KS. Complete sequence of the mitochondrial genome of Taenia saginata: comparison with T. solium and T. asiatica. Parasitol Int 2007;56(3):243-246. https://doi.org/10.1016/j.parint.2007.04.001
- 31. Li WX, Zhang D, Boyce K, Xi BW, Zou H, et al. The complete mitochondrial DNA of three monozoic tapeworms in the Caryophyllidea: a mitogenomic perspective on the phylogeny of eucestodes. Parasit Vectors 2017;10(1):314. https://doi.org/10.1186/s13071-017-2245-y
- 32. Xi BW, Oros M, Chen K, Xie J. A new monozoic tapeworm, Parabreviscolex niepini n. g., n. sp. (Cestoda: Caryophyllidea), from schizothoracine fishes (Cyprinidae: Schizothoracinae) in Tibet, China. Parasitol Res 2018;117(2):347-354. https://doi.org/10.1007/s00436-017-5682-9
- 33. Okamoto M, Bessho Y, Kamiya M, Kurosawa T, Horii T. Phylogenetic relationships within Taenia taeniaeformis variants and other taeniid cestodes inferred from the nucleotide sequence of the cytochrome c oxidase subunit I gene. Parasitol Res 1995;81(6):451-458. https://doi.org/10.1007/BF00931785
- 34. Jin YC, Li XY, Liu JH, Zhu XQ, Liu GH. Comparative analysis of mitochondrial DNA datasets indicates that Toxascaris leonina represents a species complex. Parasit Vectors 2019;12(1):194. https://doi.org/10.1186/s13071-019-3447-2
- 35. Baker RJ, Bradley RD. Speciation in mammals and the genetic species concept. J Mammal 2006;87(4):643-662. https://doi.org/10.1644/06-MAMM-F-038R2.1