Abstract
Perkinsus marinus is a major protozoan pathogen of oysters, responsible for severe mortality events and substantial economic losses in the global aquaculture industry. Rapid, sensitive, and reliable detection of this parasite is therefore essential for effective monitoring and timely control of dermo disease outbreaks. In this study, we developed and optimized a novel loop-mediated isothermal amplification (LAMP) assay, designated Pm-LAMP, for the specific detection of P. marinus in oyster tissues. The optimized Pm-LAMP assay, employing 5 primers and performed at 67°C, demonstrated high analytical sensitivity, consistently detecting DNA concentrations as low as 40 fg/µl and enabling accurate quantification down to 0.4 pg/µl. The assay exhibited linear amplification across a wide template range from 4 ng/µl to 0.4 pg/µl, with a strong inverse correlation between template concentration and threshold time. Specificity testing confirmed exclusive amplification of P. marinus, with no cross-reactivity observed for P. olseni, P. honshuensis, or P. chesapeaki. This study represents the first LAMP assay specifically designed for the detection of P. marinus. The Pm-LAMP assay was validated using Pacific oyster tissues and cultured P. marinus isolates originating from the USA and Korea and was benchmarked against quantitative real-time PCR (qPCR). Although qPCR exhibited higher sensitivity for detecting trace DNA levels, the Pm-LAMP assay produced results within 20 min while maintaining reliable detection at low DNA concentrations. Diagnostic performance evaluation showed 100% sensitivity and 90.91% specificity, with substantial agreement with qPCR (Cohen’s κ=0.811). Overall, the Pm-LAMP assay provides a rapid, robust, and field-deployable diagnostic tool for P. marinus, supporting improved disease surveillance and sustainable oyster aquaculture management.
-
Key words: Perkinsus marinus, oysters, LAMP assay, quantitative real-time PCR
Introduction
Perkinsus marinus, a protistan parasite, is the causative agent of "dermo" disease (perkinsosis), which leads to chronic infections and significant mortality in oyster populations. It poses a major threat to oyster aquaculture, particularly in the Eastern oyster (
Crassostrea virginica) along the Atlantic and Gulf coasts of the USA. Due to its high pathogenicity and economic impact,
P. marinus has been listed as a notifiable pathogen by the World Organization for Animal Health (WOAH).
P. marinus was first identified on the Gulf Coast of Mexico in the 1940s [
1], and has since been reported in various bivalve species [
2,
3], Eastern oyster (
C. virginica) is recognized as the primary host of
P. marinus [
4]. Notably,
P. marinus has also caused high mortality in Pacific oysters (
C. gigas) in the Gulf of California [
5]. More recently, its presence was confirmed in
C. gigas along the west coast of Korea, albeit at low infection intensity [
6]. The severity of
P. marinus infections typically increases during the warmer summer months, contributing to substantial economic losses in oyster-producing regions.
Traditional diagnostic methods for
P. marinus, including Ray’s fluid thioglycollate medium (RFTM) assay, histology, and conventional PCR, have been used for decades. Among these, the RFTM assay is regarded as the WOAH-recommended gold standard for
P. marinus diagnosis. Molecular methods such as conventional PCR [
7] and in situ hybridization [
8] offer improved specificity but require specialized instrumentation and laboratory workflows; by contrast, microscopy-based diagnostics (e.g., RFTM/histology) are labor-intensive and can be highly operator-dependent.
To address these limitations, quantitative real-time PCR (qPCR) has emerged as a powerful tool for sensitive and specific detection of
P. marinus DNA. It has been increasingly adopted for quantitative surveillance of this pathogen [
9,
10]. Recently, we developed an advanced qPCR assay using newly designed primers and a TaqMan-based probe with an internal quencher (i-EBQ), which significantly improves detection sensitivity and reliability [
11]. Although qPCR is highly effective, its reliance on thermocyclers and controlled laboratory conditions limits its deployment in field settings or low-resource environments.
In recent years, loop-mediated isothermal amplification (LAMP) has emerged as a promising alternative for rapid, cost-effective, and sensitive pathogen detection [
12]. Unlike PCR-based techniques, LAMP amplifies DNA at a constant temperature using a strand-displacing DNA polymerase derived from
Geobacillus stearothermophilus [
13,
14]. The LAMP reaction utilizes 4 core primers targeting 6 distinct regions of the target gene, with 2 additional loop primers to further enhance the speed and efficiency of amplification [
15,
16]. Advantages of LAMP include rapid amplification, minimal equipment requirements, and adaptability to field conditions. Amplification products can be detected via turbidity, color changes, or fluorescence, with real-time monitoring enabling quantification [
17].
LAMP assays have been successfully developed for a wide range of aquatic pathogens, including
Perkinsus spp. [
18]. Feng et al. [
19] developed a LAMP assay capable of detecting both
P. marinus and
P. olseni. Other groups have introduced species-specific LAMP assays for
P. olseni using 4 or 6 primers [
20]. A duplex LAMP assay for the simultaneous detection of
Perkinsus and
Bonamia spp. has also been reported [
21]. However, to date, no LAMP assay has been developed to exclusively detect
P. marinus, without cross-amplifying other
Perkinsus species.
Here, we report the development and validation of a Pm-LAMP, species-specific LAMP assay targeting a P. marinus hypothetical-protein locus. We describe primer design and in silico specificity screening, optimization of isothermal conditions and primer combinations, and analytical evaluations (limit of detection and exclusivity) alongside diagnostic performance benchmarking against qPCR using cultured strains and oyster tissues from Korea and the USA. Our goal was to establish a rapid, instrument-lean workflow suitable for routine surveillance of P. marinus in aquaculture settings.
Methods
Ethics statement
Not applicable.
Tissue samples, cells, and DNA extraction
Pacific oyster samples (
n=16), collected from the West Coast of Korea, were used as environmental samples in this study. Additionally, 6
P. marinus strains from different locations in the United States were obtained from the American Type Culture Collection (ATCC), with the following ATCC numbers: 50894, 50509, 50510, 50787, 50766, and 50849. Two other
Perkinsus species,
P. honshuensis (ATCC PRA-176) and
P. chesapeaki (ATCC PRA-65), were also sourced from ATCC.
P. olseni, originally isolated from Manila clams (
Ruditapes philippinarum) in our laboratory [
22], was also included in the experiments. All cells were maintained in standard DMEM/F-12 medium following previously established protocols [
22,
23]. Total DNA was extracted from approximately 25 mg of mantle tissue or cultured cells using a commercial DNA extraction kit (Qiagen) following the manufacturer’s protocol. The extracted DNA was stored at -20°C until further use.
The presence of
P. marinus in the mantle tissues of Pacific oysters and the 6 strains obtained from ATCC was confirmed via conventional PCR using species-specific primers developed by Audemard et al. [
7] and recommended by World Organization for Animal Health [
24] (
Table 1) [
11,
16,
25]. The PCR reaction was carried out in a 50-µl mixture containing 0.4 µM of each primer (forward: PmarITS-70F and reverse: PmarITS600R), 0.2 mM dNTP mix, 10× Taq reaction Buffer (SolGent), 1.25 U of Solg Taq DNA polymerase (SolGent), 5 µl of template DNA, and nuclease-free water to final volume 50 µl. PCR amplification was performed using a SimpliAmp Thermal cycler (Thermo Fisher Scientific) under the following cycling conditions: initial denaturation at 95°C for 4 min; 40 cycles of denaturation at 94°C for 1 min; annealing at 57°C for 1 min; elongation at 65°C for 3 min; followed by a final elongation at 65°C for 10 min. PCR products were analyzed by electrophoresis on a 1.5% agarose gel.
The qPCR assay for
P. marinus (Pm-qPCR) was conducted using a recently developed assay [
11]. Briefly, the 25-µl reaction mixture included 0.6 µM of each primer and 0.4 µM of probes targeting the hypothetical protein gene of
P. marinus and the movement protein gene of the tobacco mosaic virus (used as an internal control, IC). The mixture also included 15 µl of AccuPower Dual-HotStart RT-qPCR Master Mix (Bioneer), 5 µl of sample DNA, and 1 µl of IC template DNA. Because the template was genomic DNA, no reverse-transcription step was performed; reactions were run as DNA qPCR using the manufacturer’s Dual-HotStart mix. Amplification was performed on a CFX Opus 96 Real-Time PCR System (Bio-Rad Laboratories) with the following cycling conditions: 95°C for 5 min, then 45 cycles of 95°C for 10 sec and 55°C for 20 sec. Fluorescence was acquired on the FAM channel for
P. marinus and the Cy5 channel for the IC. Result interpretation followed our prior validation [
11]: positive if Ct ≤35.8 with a sigmoidal curve and valid IC; Negative if Ct >35.8 or no amplification by 45 cycles with a valid IC.
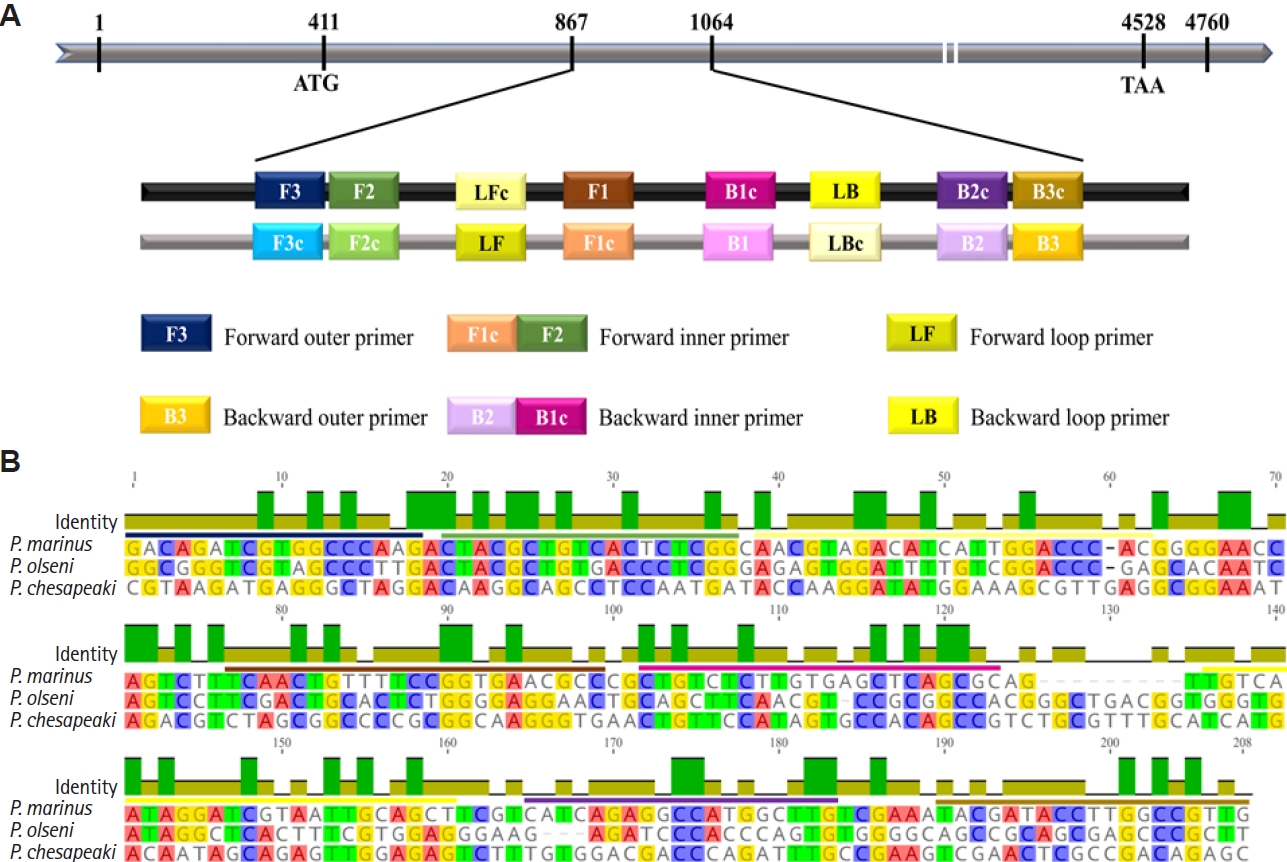
LAMP primers were designed using the online primer design tool developed by New England Biolabs (NEB,
https://lamp.neb.com) with default parameters. A genomic DNA sequence coding for a hypothetical protein was selected from the whole-genome sequence database of
P. marinus (GenBank accession No. NW_003206658.1, ATCC 50983 genomic scaffold scf_1104296958712) as the target gene. A total of 6 primers, including 2 outer (forward outer and backward outer), 2 inner (forward inner and backward inner), and 2 loop primers (loop forward [LF] and loop backward [LB]), were designed to recognize 8 distinct regions on the target gene or its complementary sequence (
Table 1;
Fig. 1A). To verify primer specificity, multiple sequence alignment was performed against homologous gene sequences of
P. olseni and
P. chesapeaki retrieved from NCBI GenBank database. The alignments were conducted using the MUSCLE algorithm within the Geneious Prime software version 2023.1.2 (Geneious).
A 504-bp genomic DNA fragment of the target gene, encompassing the regions recognized by the designed LAMP primers, was custom-synthesized by Macrogen. The synthetic DNA fragment was subsequently cloned into the pMG-Amp vector (Macrogen) to serve as a standard template for the Pm-LAMP assay.
Real-time fluorometric Pm-LAMP assay setup
The Pm-LAMP reactions were carried out using a WarmStart LAMP kit (E1700L, NEB) designed for real-time fluorescence detection. The Pm-LAMP was conducted in a 20-µl reaction mixture containing 10 µl of WarmStart LAMP 2× Master Mix (NEB), 1.6 µM of each inner primer, 0.2 µM each outer primer, 0.4 µM each loop primers, 0.5 µl of 50× fluorescence dye, and either 1 µl of standard plasmid DNA or 5 µl of sample template DNA. PCR-grade water was added to bring the final volume to 20 µl. No-template controls (NTCs) included PCR-grade water instead of DNA to ensure the absence of non-specific amplification.
Amplification was performed isothermally at 65°C on a CFX Opus 96 Real-Time PCR detection system (Bio-Rad Laboratories) to record threshold-time (Tt) values and enable direct comparison with qPCR during assay optimization. Fluorescence was acquired every 30 sec on the FAM channel over a 30-min incubation at 65°C. For specificity confirmation, a post-amplification melting curve analysis was run from 65°C to 95°C in 0.5°C increments every 5 sec.
Pm-LAMP assay optimization
The optimal temperature for the Pm-LAMP assay was initially determined by conducting amplification reactions at temperatures ranging from 55°C to 70°C. Based on the results, 67°C was identified as optimal for subsequent experiments. To address non-specific amplification observed in the NTCs, additional optimization was conducted by modifying the standard Pm-LAMP protocol described in Section 2.5. Specifically, reactions were performed using 5 primers by excluding either the LF or the LB primers to assess their impact on non-specific amplification in NTCs. Amplification curves and melting peaks were analyzed, and Tt and melting temperature (Tm) values were recorded to evaluate the effectiveness and specificity of each reaction condition. After selecting the final configuration (5 primers, 67°C, LF omitted), we re-measured Tt and Tm; unless otherwise noted, all Tt and Tm values reported in the results refer to this finalized assay.
Sensitivity of Pm-LAMP assay
The sensitivity of the optimized Pm-LAMP assay, which uses a 5-primer system including the LB primer, was evaluated using 10-fold serial dilutions of standard P. marinus template DNA, with concentrations ranging from 4 ng/µl to 0.4 fg/µl. All reactions were conducted at 67°C using a real-time PCR system, and melting curve analysis was performed to confirm amplification specificity. Tt values obtained from each dilution were used to generate a standard curve, allowing the quantification of assay performance and determination of the detection limit.
Specificity of Pm-LAMP assay
The specificity of the optimized Pm-LAMP assay was tested using total DNA (approximately 50 ng/µl) extracted from P. honshuensis (ATCC PRA-176), P. chesapeaki (ATCC PRA-65), and P. olseni. This assay ensured that the Pm-LAMP assay exclusively detected P. marinus without cross-reactivity with other Perkinsus species.
Validation of Pm-LAMP assay
The Pm-LAMP assay was validated using tissue samples from 16 Pacific oysters collected from the West Coast of Korea, as well as 6
P. marinus strains obtained from ATCC. Additionally, a Korean
P. marinus strain isolated from Pacific oysters [
6] was included in the validation process. A 10-fold dilution series of the Korean
P. marinus sample (56 ng/µl) was tested using both Pm-LAMP and qPCR assays. The optimized Pm-LAMP assay was employed to determine the Tt for each sample by measuring the fluorescence intensities of the amplified targets. Specific amplification was confirmed by evaluating the melting peaks. The results from the Pm-LAMP assay were then compared with those obtained from conventional PCR and/or qPCR to assess performance and reliability.
To validate the results, both amplification plots and melting peaks were thoroughly examined. Standard curve for the test was generated using Excel 2019 (Microsoft), and the coefficient of determination (R²) was calculated to assess the quantification capability of the Pm-LAMP assay. For the optimized Pm-LAMP assay, clinical diagnostic sensitivity and specificity were estimated using a confusion matrix (2×2 contingency table) [
26]. These metrics were compared against results from a recently developed qPCR assay [
11]. In this analysis, samples that tested positive by both qPCR and LAMP were classified as true positives, while those that tested negative by both methods were classified as true negatives. Samples that were positive by LAMP but negative by qPCR were considered as false positives.
Cohen’s kappa test was performed to assess the reliability of the optimized Pm-LAMP assay. Cohen’s kappa coefficient (κ) was calculated using the GraphPad online statistical tool (
https://www.graphpad.com/quickcalcs/kappa1/) with a 95% confidence interval. The κ-value ranges from 0 (indicating no agreement) to 1 (indicating perfect agreement). The κ-values were interpreted as follows: perfect agreement (0.81–1.00), substantial agreement (0.61–0.80), moderate agreement (0.41–0.60), and fair agreement (0.21–0.40).
Results
Development of specific LAMP primers for P. marinus
Multiple sequence alignment of the genomic DNA sequence of a hypothetical protein of
P. marinus with corresponding regions of
P. olseni and
P. chesapeaki revealed a 198-bp region with high interspecific variations. Six specific LAMP primers were designed within this region, as depicted in
Fig. 1B.
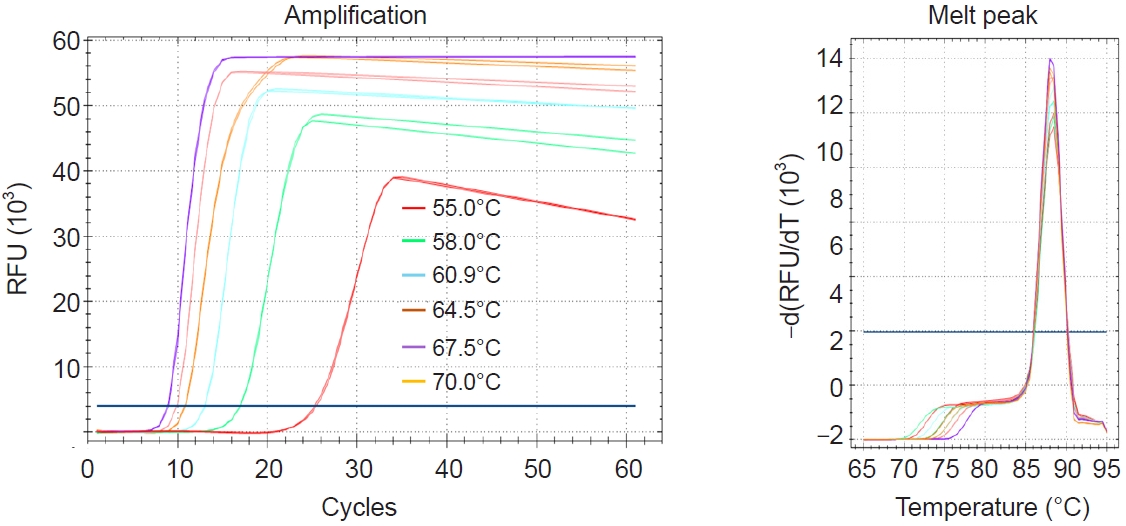
Temperature
The initial LAMP assay was performed at various temperatures ranging from 55°C to 70°C using 6 primers. The results revealed that the lowest Tt value was observed at 67.5°C (
Fig. 2). Therefore, 67°C was selected as the optimal reaction temperature for the LAMP primers designed in this study, and all subsequent reactions were conducted at this temperature.
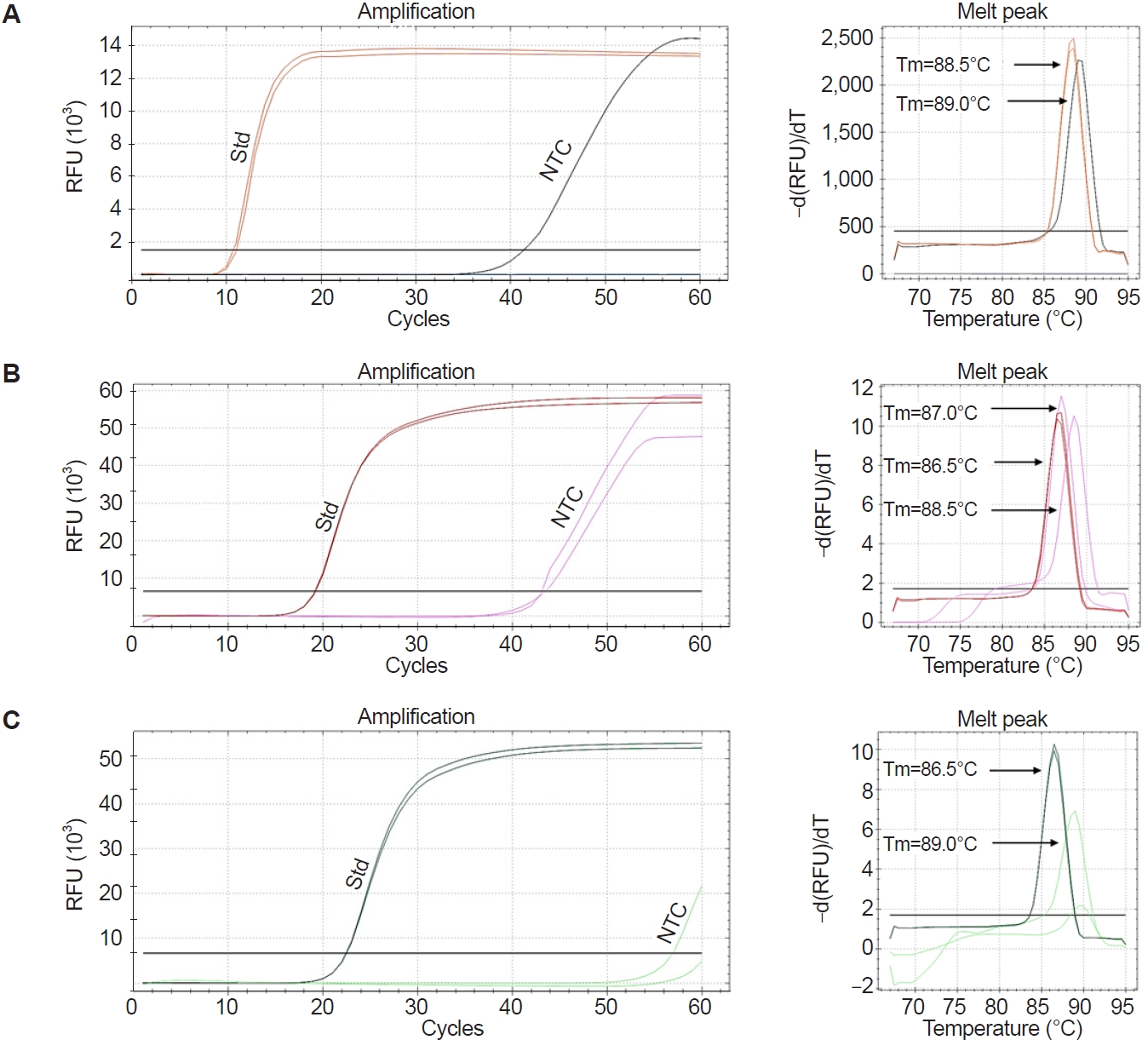
Five primers
During optimization at 67°C, the LAMP assay exhibited non-specific amplification in the NTC, as shown in
Fig. 3A. To further refine the assay, 5-primer reactions were tested by excluding either the LF or LB primer. These reactions also produced non-specific amplification in the NTCs. Notably, when the LB primer was used (i.e., LF excluded), amplification in the NTCs was observed occasionally only after 50 cycles (
Fig. 3B,
C), suggesting improved specificity compared to other conditions. Further, distinct melting peaks were observed in the specific and non-specific amplifications. The Tm values for standards ranged from 86.5°C to 87.0°C, while those for the NTCs ranged from 88.5°C to 89.0°C, particularly when the LF primer was excluded (
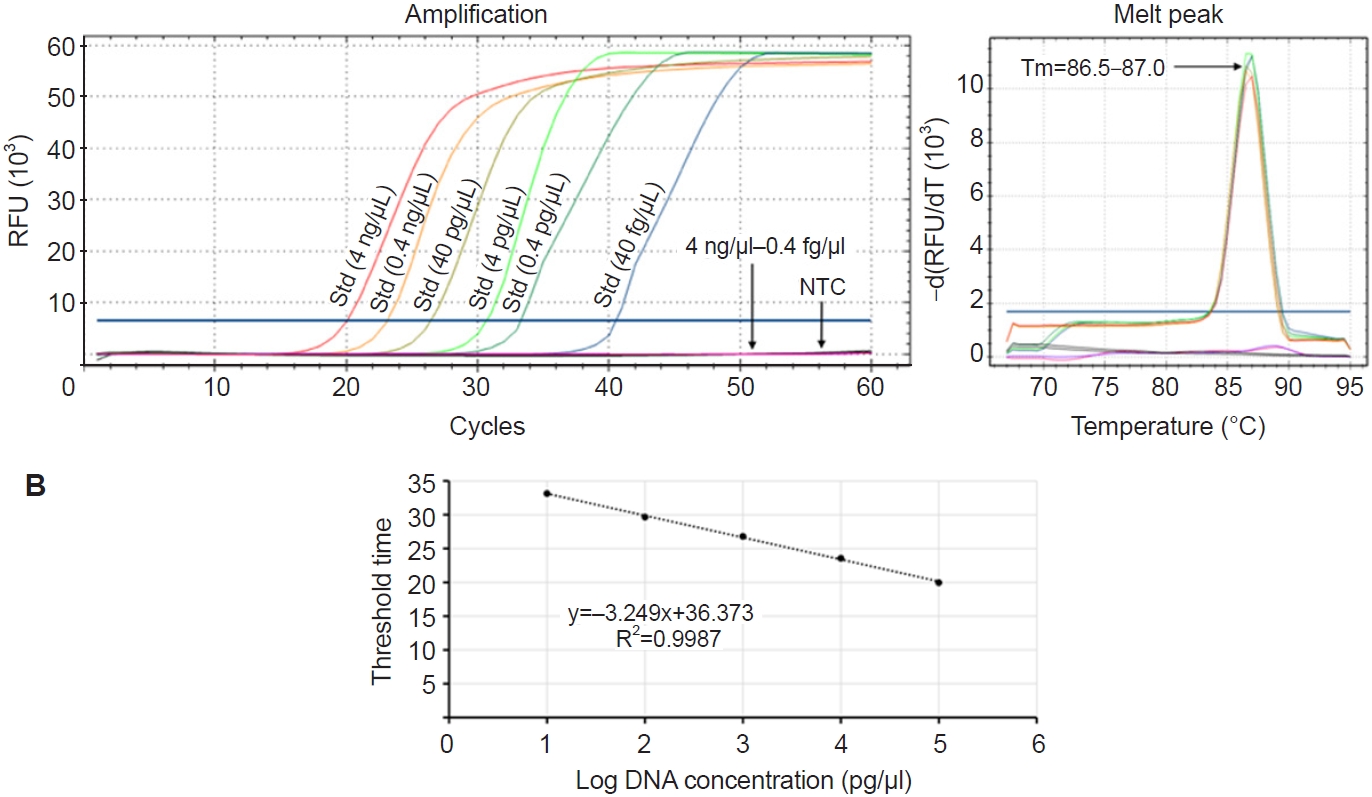
Fig. 3C). Based on these observations, the LAMP assay using the LB (without LF) was selected as the optimized method for detecting
P. marinus. This optimized assay showed linear amplification across template concentrations from 4 ng/µl to 0.4 pg/µl (
Fig. 4A), with a strong correlation between template concentration and Tt values (
R2 = 0.9987) (
Fig. 4B). According to
Fig. 4A, 0.4 pg/µl—the lowest concentration that could be accurately quantified—was amplified in under 34 cycles, which corresponds to approximately 17 min. This indicates that the LAMP assay is not only sensitive and quantitative but also rapid, making it suitable for timely detection of
P. marinus.
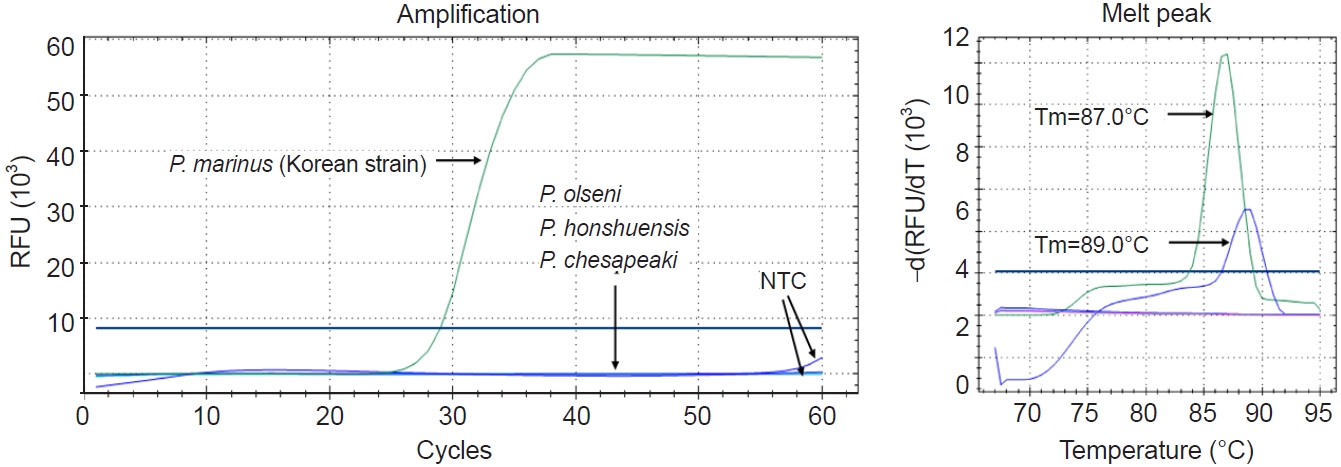
Three species,
P. olseni,
P. honshuensis, and
P. chesapeaki, were tested to assess the analytical specificity of the Pm-LAMP assay developed herein. No specific amplification was observed for any of these species, confirming the high specificity of the assay for
P. marinus (
Fig. 5;
Table 2).
The Pm-LAMP assay was successfully applied to 6
P. marinus strains and cultured cells isolated from the West Coast of Korea. Amplification was observed in all cases, and the specific product exhibited a single melting curve within 86.5°C–87.0°C (
Table 2). Among 16 environmental oyster tissue samples, 5 tested positive for
P. marinus using conventional PCR, as confirmed by agarose gel electrophoresis (data not shown). Using the validated cutoff from a previous study [
11], Pm-qPCR calls were positive if Ct ≤ 35.8 and negative if Ct >35.8 (with valid IC and appropriate curve shape). By Pm-qPCR, 7 samples were positive (high Ct values) and 9 were negative. The optimized Pm-LAMP detected 6 positives that met the melting curve criterion above, while 8 samples tested negative. Two additional samples produced amplification with a single peak in the 86.5°C–87.0°C window but were classified as false positives for diagnostic analysis because both conventional PCR and Pm-qPCR were negative (
Table 3). Sample No. 7 was positive by qPCR (Ct ≤35.8) but negative by Pm-LAMP owing to a very late Tt and a Tm of 88.5°C, which falls outside the 86.5°C–87.0°C acceptance window; thus, it was adjudicated as discordant. Analysis of DNA extracted from
P. marinus cells indicated that both the Pm-qPCR and Pm-LAMP assays could detect
P. marinus at concentrations as low as 56 pg/µl (
Table 4). The clinical diagnostic sensitivity and diagnostic specificity of the Pm-LAMP assay for detecting
P. marinus were 100% and 80.0%, respectively. Cohen’s kappa analysis confirmed substantial agreement between the Pm-LAMP and Pm-qPCR assays, with a κ-value of 0.750.
Discussion
P. marinus poses a significant threat to oyster populations, causing substantial mortality and economic losses in the aquaculture industry [
5,
27]. Rapid detection of
P. marinus is essential for effectively managing and controlling dermo disease outbreaks in oyster farms. Molecular techniques, such as conventional PCR [
7] and qPCR [
9,
10], are widely used; indeed, we recently developed a highly specific and sensitive qPCR assay for
P. marinus [
11]. However, these approaches require specialized instruments and longer turnaround times. LAMP assay offers a valuable alternative by enabling rapid and specific DNA amplification under isothermal conditions. In this study, we developed and validated a
P. marinus-specific LAMP assay for rapid, quantitative detection of the parasite, and we benchmarked its performance against our previously established Pm-qPCR [
11]. The results demonstrate the assay’s potential as a robust diagnostic tool for
P. marinus.
For analytical benchmarking, LAMP Tt were recorded on a laboratory real-time platform to standardize optimization and allow head-to-head comparison with qPCR. This choice does not constrain deployment: the same primers and chemistry are compatible with simple constant-temperature heaters and endpoint visual/fluorometric readouts suitable for field or low-resource settings.
We selected a species-specific target, an uncharacterized (hypothetical-protein) locus from the
P. marinus whole-genome sequence database available in the NCBI. Homology analysis confirmed low sequence conservation between this target sequence and the corresponding regions in
P. olseni and
P. chesapeaki, supporting the high specificity of the designed primers. Previous LAMP assays for
Perkinsus spp. primarily targeted the ITS region, which is not species-specific for
P. marinus [
19,
21]. In contrast, the target used here preserved species-level exclusivity without major loss of analytical sensitivity: Pm-LAMP detected cultured
P. marinus DNA down to 56 pg/µl and yielded faster time-to-result than qPCR. Two LAMP-positive/qPCR-negative environmental samples exhibited late Tt yet a single melting curve within the acceptance window; we classified these as false positives for diagnostic analysis because neither conventional PCR nor Pm-qPCR confirmed them. Such borderline, late-appearing signals can occur at very low template input; conservative Tt cutoffs, carryover-prevention chemistry (UDG/dUTP), and—where available—portable melting curve checks help minimize their occurrence.
Optimization of the LAMP protocol revealed that non-specific amplification occurred in NTCs at the initial temperature of 65°C. We evaluated higher reaction temperatures (up to 70°C), since previous studies noted that elevated temperatures can improve specificity and reduce non-specific amplification [
28,
29]. At 67°C, the assay yielded the lowest Tt value and a marked reduction in non-specific amplification, identifying 67°C as the optimal amplification temperature. In addition, to further minimize background amplification, the LF primer was removed, based on recent reports of similar LAMP optimization [
30]. This modification significantly reduced non-specific amplification. The fully optimized Pm-LAMP assay (employing 5 primers at 67°C) exhibited high sensitivity, consistently detecting as little as 40 fg/µl of
P. marinus DNA and accurately quantifying down to 0.4 pg/µl.
The analytical specificity of the assay was confirmed using genomic DNA from P. olseni, P. honshuensis, and P. chesapeaki—none of these non-target species yielded amplification, whereas amplification was observed exclusively for P. marinus. In environmental oyster tissue samples, Pm-LAMP showed high analytical specificity (single melting curve within 86.5°C–87.0°C) and good diagnostic agreement with qPCR (diagnostic sensitivity=100%, diagnostic specificity=80.0%, κ=0.750). While qPCR was more sensitive for trace DNA, Pm-LAMP delivered substantially faster results, supporting its use as a practical alternative with appropriate confirmatory safeguards.
Given that the primary host of
P. marinus is the Eastern oyster (
C. virginica) [
4], the Pm-LAMP assay could be especially beneficial in regions where
C. virginica is cultured by enabling rapid on-site detection of
P. marinus. Notably, in our study, the Pm-LAMP assay could detect as little as 40 fg/µl of target DNA with a Tt of approximately 20 min. In contrast, a 0.4 pg/µl template produced a positive LAMP result in about 17 min. These rapid detection times, even at very low DNA concentrations, underscore the assay’s efficiency for quick diagnostics in the field.
Operationally, most non-specific LAMP products were filtered by melting curve analysis: true positives exhibited a single peak within 86.5°C–87.0°C, whereas non-specific signals showed higher Tm values (≈88.5°C–89.0°C). To prioritize specificity, we apply a conservative Tt cutoff, scoring late signals as indeterminate and retesting; in our cohort, 2 LAMP-positive/qPCR-negative samples met the Tm window but had late Tt and were classified as false positives for diagnostic analysis. Melting curve analysis is widely used to differentiate LAMP products among parasite species or gene targets [
31,
32]. For field use, portable fluorescence LAMP analyzers can run a brief post-amplification melting curve analysis step to verify the expected Tm (≈86.5°C–87.0°C); where such devices are unavailable, instrument-free strategies are viable, including closed-tube colorimetric LAMP, lateral-flow dipstick confirmation, and procedural safeguards (conservative Tt cutoffs and UDG/dUTP carryover prevention).
Field applicability is further supported by portable isothermal fluorescence devices (e.g., Genie-class instruments such as Genie II, OptiGene) that provide real-time detection without conventional thermal cycling and can optionally run a brief melting curve check [
25]. In this study, all oyster tissue DNAs were purified using a commercial kit; to fully enable field deployment, future work will evaluate simplified extraction (e.g., heat treatment or alkaline lysis) and perform head-to-head Pm-LAMP vs. qPCR comparisons on crude extracts for low-burden samples, building on field-deployable LAMP workflows [
33-
35]. Integrating such extraction-light methods with portable platforms should allow on-site testing by non-experts, reducing cost and infrastructure needs while facilitating real-time surveillance of
P. marinus in aquaculture settings.
In summary, the Pm-LAMP assay developed here is a robust, efficient method for P. marinus detection that complements existing qPCR workflows. Optimized at 67°C in a 5-primer configuration (LF omitted), the assay showed high analytical specificity and good diagnostic agreement with qPCR. Ongoing work will focus on enhancing sensitivity for low-burden samples, extending the assay to environmental DNA monitoring, and validating extraction-light, field-friendly confirmation options (e.g., portable anneal/melt checks or lateral-flow dipsticks), thereby broadening the assay’s impact across diagnostic and ecological settings.
Notes
-
Author contributions
Conceptualization: Bathige SDNK. Funding acquisition: Park KI. Methodology: Bathige SDNK, Kim SH, Lee D, Jeon HB. Project administration: Park KI. Resources: Chen Y. Supervision: Park KI. Writing – original draft: Bathige SDNK. Writing – review & editing: Bathige SDNK, Jeon HB, Chen Y, Park KI.
-
Conflict of interest
The authors have no conflicts of interest to declare.
-
Funding
This study was supported by the Korea Institute of Marine Science & Technology Promotion (KIMST), funded by the Ministry of Oceans and Fisheries, Korea (grant No. RS-2022-KS221679) and the National Research Foundation of Korea (grant No. RS-2022-NR069484).
Fig. 1.Design of loop-mediated isothermal amplification (LAMP) primers specific for Perkinsus marinus based on the genomic sequence of a hypothetical protein. (A) Schematic representation of primers designed for the LAMP assay. Numbers in the grey bar represent the nucleotide positions. (B) Multiple sequence alignment performed using the MUSCLE algorithm, comparing with P. marinus and 2 related species, P. olseni and P. chesapeaki. Colored lines above the sequence indicate the designed primer regions, with sequence identity represented by a green bar.
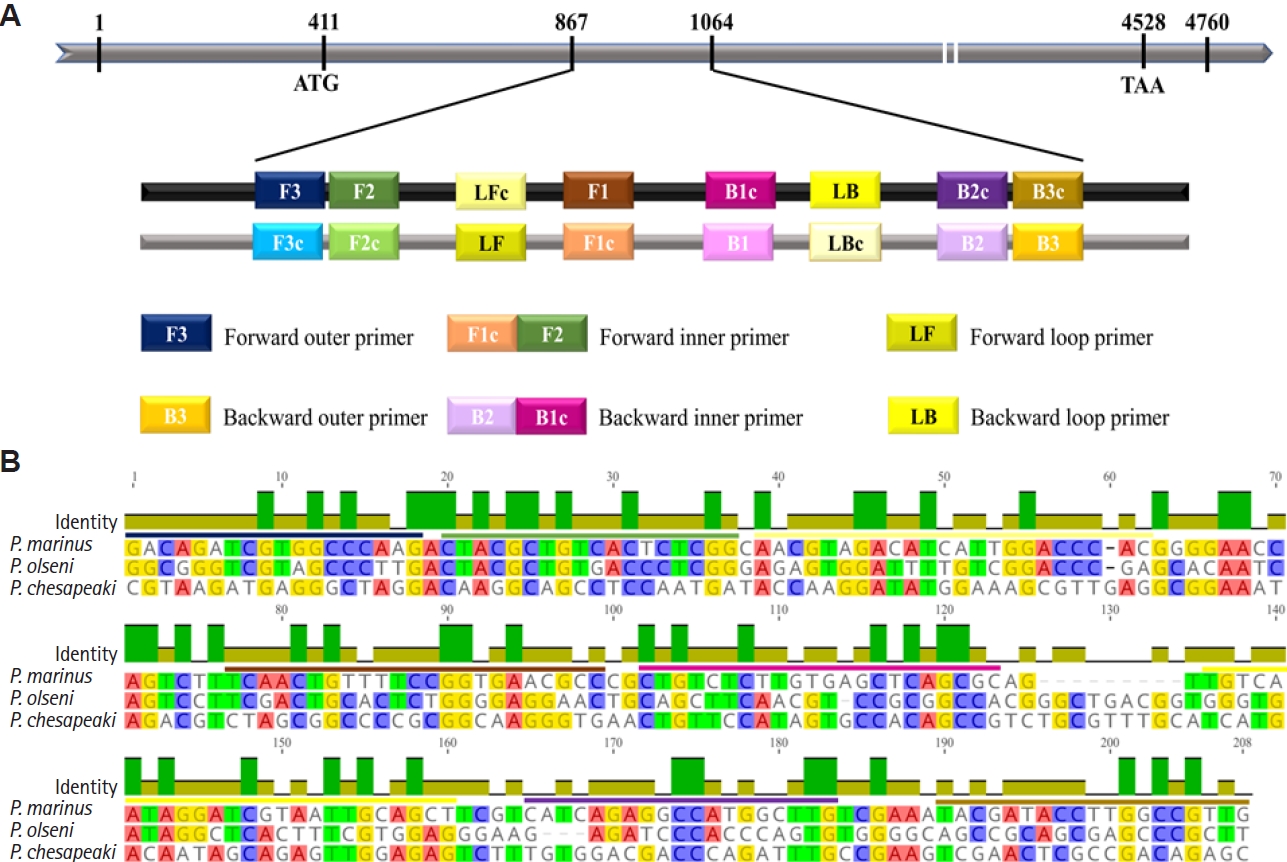
Fig. 2.Evaluation of optimum temperature for the Pm-LAMP (species-specific loop-mediated isothermal amplification assay targeting a Perkinsus marinus hypothetical-protein locus) assay. Amplification plots and melting peaks at different temperatures from 55C to 70C were assessed using temperature gradient. RFU, relative fluorescence units.
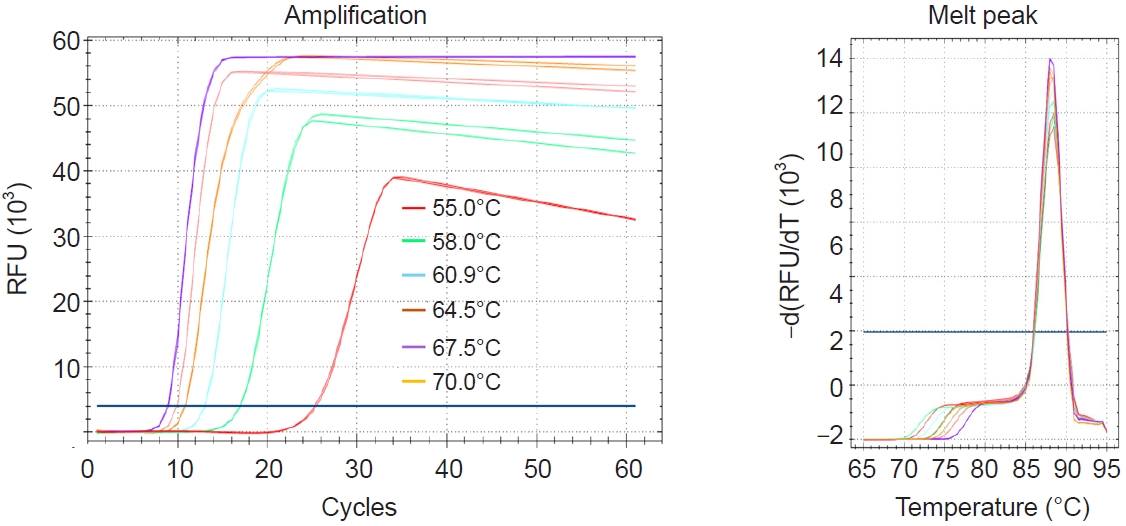
Fig. 3.Evaluation of Pm-LAMP (species-specific loop-mediated isothermal amplification [LAMP] assay targeting a Perkinsus marinus hypothetical-protein locus) with a combination of LAMP primers. Amplification plots and melting peaks for reaction with (A) 6 primers, (B) 5 primers (without loop backward) and (C) 5 primers (without loop forward). Amplification plots for the P. marinus standard (Std) and no template control (NTC) were marked. The melting temperature (Tm) of each melting peak of the standard and NTC is displayed. RFU, relative fluorescence units.
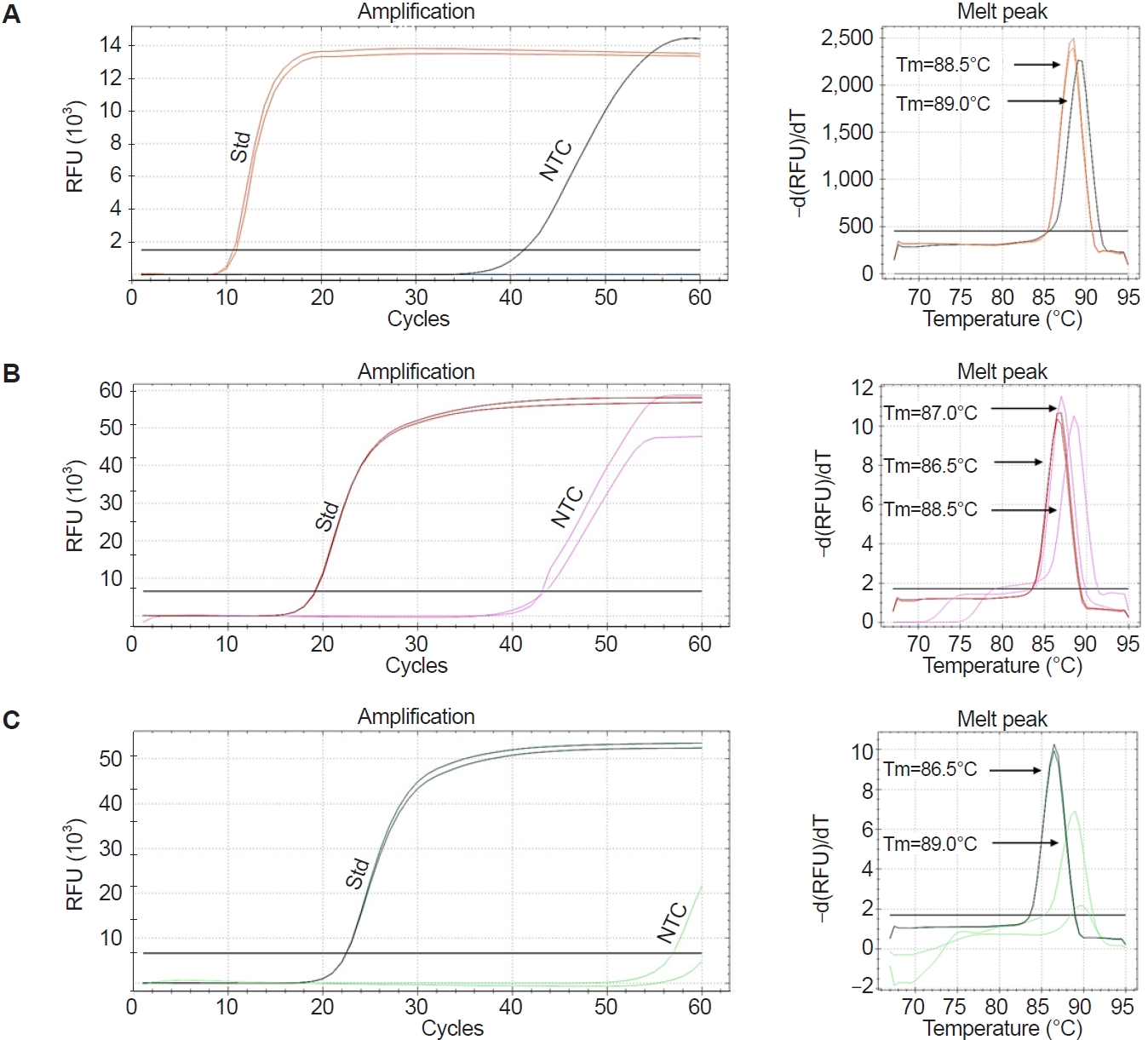
Fig. 4.Evaluation of limit of detection. (A) Amplification plots for a 10-fold dilution series of standards (4 ng/µl–0.4 fg/µl) without the loop forward primer. (B) Standard curve generated based on the threshold time and log value of corresponding template concentrations. RFU, relative fluorescence units; Std, standard; NTC, no template control.
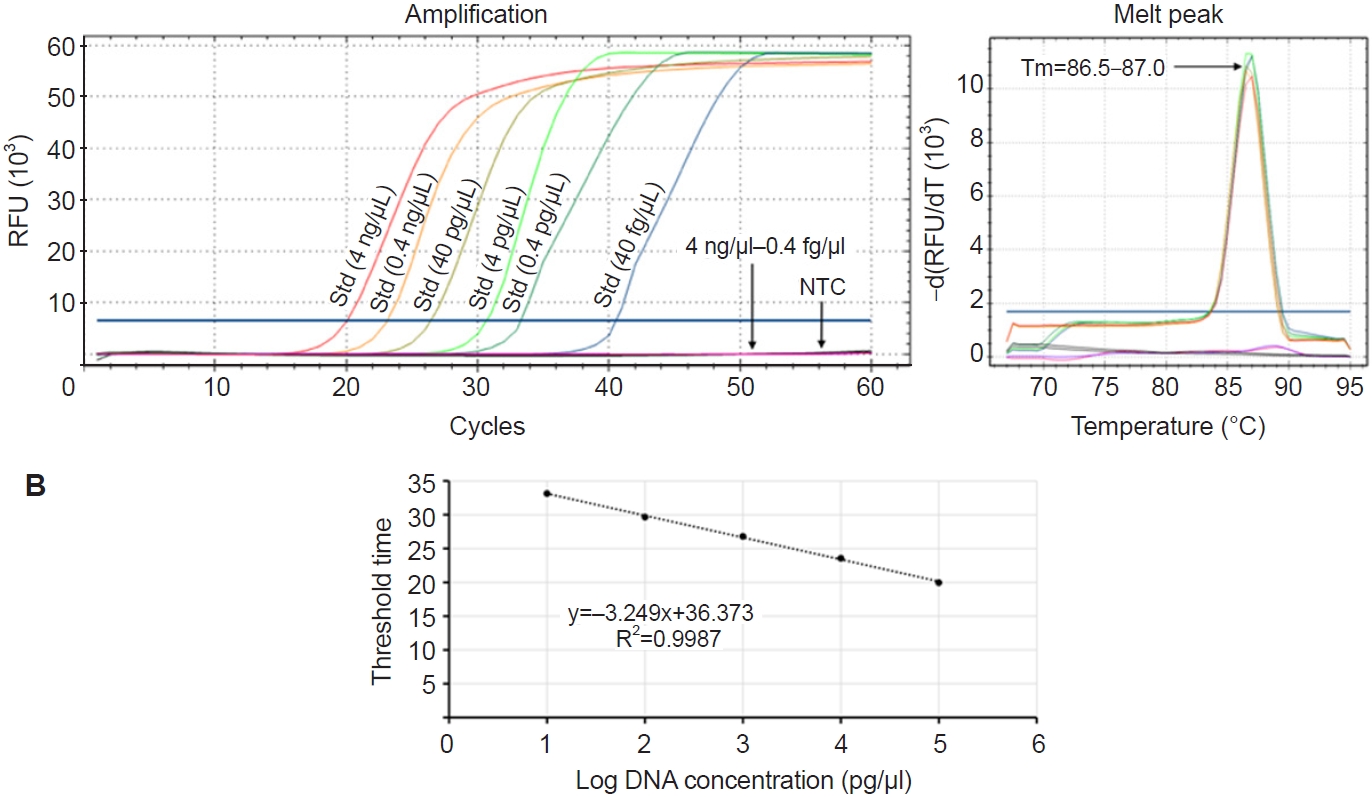
Fig. 5.Evaluation of the analytical specificity of Pm-LAMP (species-specific loop-mediated isothermal amplification assay targeting a Perkinsus marinus hypothetical-protein locus) assay. Amplification plots and melting peaks for P. marinus and 3 other species: P. olseni, P. honshuensis, and P. chesapeaki. Two distinct melting peaks were observed for the P. marinus standard and no template control (NTC), each with different melting temperature (Tm) values. RFU, relative fluorescence units.
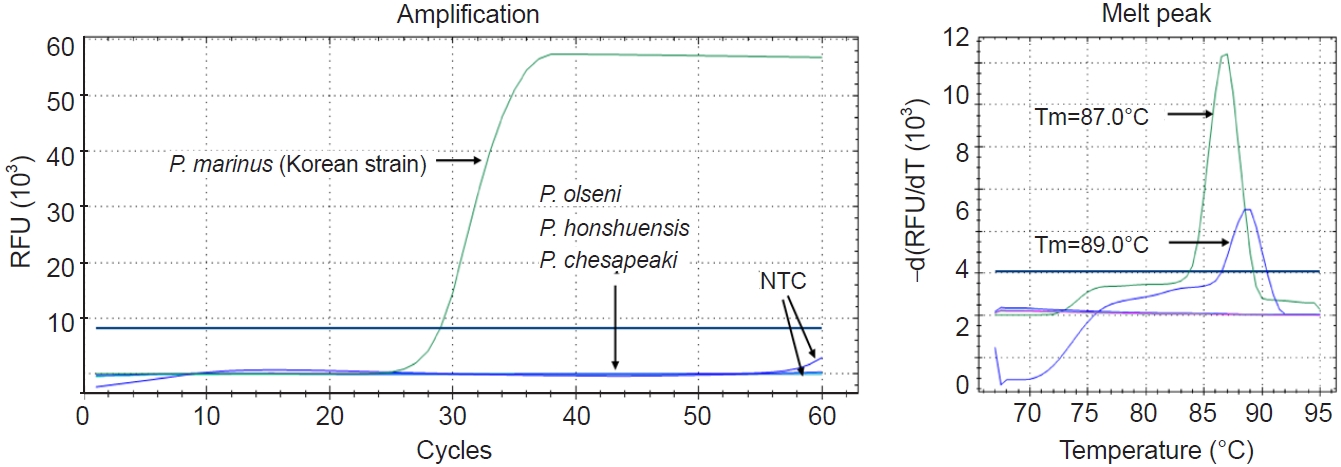
Table 1.Primers used in this study
Table 1.
|
Experiment |
Primer name |
Primer sequence (5′→3′) |
Target gene |
Reference |
|
PCR |
PmarITS-70F |
CTTTTGYTWGAGWGTTGCGAGATG |
ITS |
[11,25] |
|
PmarITS600R |
CGAGTTTGCGAGTACCTCKAGAG |
|
qPCR |
PmHP-F |
CCCAGTTCACAGTGCCTGTC |
Hypothetical protein |
[16] |
|
PmHP-R |
CATGGAATGCCGAGGGTACA |
|
PmHP-P |
[FAM]AGCGTCAT[i-EBQ]CGGACCTCGTGCA[Phosphate] |
|
LAMP |
PmF3 |
GACAGATCGTGGCCCAAG |
Hypothetical protein |
This study |
|
PmB3 |
CAACGGCCAAGGTATCGTAT |
|
PmFIP |
GGCGTTCACCGGAAAACAGTTGACTACGCTGTCACTCTCGG |
|
PmBIP |
CTGTCTCTTGTGAGCTCAGCGCCAAGCCATGGCCTCTGATG |
|
PmLF |
GTGGGTCCAATGATGTCTACGTT |
|
PmLB |
TGTCAATAGGATCGTAATTGCAGCT |
Table 2.Pm-LAMP performance on cultured isolates and non-target species
Table 2.
|
Species |
Source |
Pm-LAMP |
Results |
|
Tt |
Tm |
|
P. marinus
|
ATCC 50894 |
33.0 |
86.5 |
Positive |
|
P. marinus
|
ATCC 50509 |
34.5 |
86.5 |
Positive |
|
P. marinus
|
ATCC 50510 |
32.6 |
87.0 |
Positive |
|
P. marinus
|
ATCC 50787 |
27.7 |
87.0 |
Positive |
|
P. marinus
|
ATCC 50766 |
28.0 |
87.0 |
Positive |
|
P. marinus
|
ATCC 50849 |
32.1 |
87.0 |
Positive |
|
P. marinus
|
Korea |
28.6 |
87.0 |
Positive |
|
P. olseni
|
Korea |
ND |
None |
Negative |
|
P. honshuensis
|
ATCC PRA-176 |
ND |
None |
Negative |
|
P. chesapeaki
|
ATCC PRA-65 |
ND |
None |
Negative |
Table 3.Summary of environmental oyster tissue samples analyzed by the Pm-LAMP assay with comparison to conventional PCR and Pm-qPCR results
Table 3.
|
Sample No. |
PCR |
qPCR |
LAMP |
|
Ct |
Interpretation |
Tt |
Tm |
Interpretation |
|
1 |
P |
35.2 |
P |
38.3 |
86.5 |
P |
|
2 |
P |
31.5 |
P |
37.4 |
87.0 |
P |
|
3 |
P |
33.2 |
P |
40.5 |
86.5 |
P |
|
4 |
P |
31.7 |
P |
47.2 |
87.0 |
P |
|
5 |
P |
34.8 |
P |
43.5 |
87.0 |
P |
|
6 |
ND |
35.2 |
P |
42.4 |
87.0 |
P |
|
7 |
ND |
34.1 |
P |
50.9 |
88.5 |
ND |
|
8 |
ND |
36.3 |
ND |
32.0a
|
87.0a
|
Pa
|
|
9 |
ND |
36.2 |
ND |
41.8a
|
87.0a
|
Pa
|
|
10 |
ND |
36.8 |
ND |
46.3 |
88.5 |
ND |
|
11 |
ND |
36.7 |
ND |
46.8 |
88.5 |
ND |
|
12 |
ND |
36.8 |
ND |
ND |
None |
ND |
|
13 |
ND |
36.0 |
ND |
49.7 |
88.5 |
ND |
|
14 |
ND |
36.0 |
ND |
ND |
None |
ND |
|
15 |
ND |
37.0 |
ND |
42.2 |
89.0 |
ND |
|
16 |
ND |
36.2 |
ND |
42.7 |
89.0 |
ND |
Table 4.Summary of 10-fold dilution series analysis of a Perkinsus marinus Korean strain using the Pm-LAMP assay and comparison with the results Pm-qPCR method
Table 4.
|
Sample dilution |
DNA concentration |
qPCR |
LAMP |
|
Ct |
Interpretation |
Tt |
Tm |
Interpretation |
|
1 |
56 ng/µl |
22.6 |
P |
27.7 |
86.5 |
P |
|
1/10 |
5.6 ng/µl |
26.0 |
P |
29.1 |
87.0 |
P |
|
1/100 |
0.56 ng/µl |
29.3 |
P |
32.3 |
87.0 |
P |
|
1/1000 |
56 pg/µl |
32.9 |
P |
42.5 |
87.0 |
P |
|
1/10000 |
5.6 pg/µl |
36.6 |
ND |
ND |
None |
ND |
|
NTC |
NTC |
ND |
ND |
ND |
None |
ND |
References
- 1. Ray SM. Biological studies of Dermocystidium marinum, a fungus parasite of oysters. Rice University; 1954.
- 2. Cáceres-Martínez J, Vásquez-Yeomans R, Padilla-Lardizábal G. Parasites of the pleasure oyster Crassostrea corteziensis cultured in Nayarit, Mexico. J Aquat Anim Health 2010;22:141-51. https://doi.org/10.1577/H09-052.1
- 3. Cáceres-Martínez J, Vásquez-Yeomans R, Padilla-Lardizábal G, del Río Portilla MA. Perkinsus marinus in pleasure oyster Crassostrea corteziensis from Nayarit, Pacific coast of México. J Invertebr Pathol 2008;99:66-73. https://doi.org/10.1016/j.jip.2008.03.005
- 4. Andrews JD. Epizootiology of the disease caused by the oyster pathogen Perkinsus marinus and its effects on the oyster industry. Am Fish Soc Spec Publ 1988;18:47-63.
- 5. Enríquez-Espinoza TL, Grijalva-Chon JM, Castro-Longoria R, Ramos-Paredes J. Perkinsus marinus in Crassostrea gigas in the Gulf of California. Dis Aquat Organ 2010;89:269-73. https://doi.org/10.3354/dao02199
- 6. Kim SH, Bathige SDNK, Jeon HB, et al. First report of Perkinsus marinus occurrence associated with wild Pacific oysters Crassostrea gigas from the west coast of Korea. J Invertebr Pathol 2024;204:108119. https://doi.org/10.1016/j.jip.2024.108119
- 7. Audemard C, Reece KS, Burreson EM. Real-time PCR for detection and quantification of the protistan parasite Perkinsus marinus in environmental waters. Appl Environ Microbiol 2004;70:6611-8. https://doi.org/10.1128/AEM.70.11.6611-6618.2004
- 8. Moss JA, Burreson EM, Reece KS. Advanced Perkinsus marinus infections in Crassostrea ariakensis maintained under laboratory conditions. J Shellfish Res 2006;25:65-72. https://doi.org/10.2983/0730-8000(2006)25[65:APMIIC]2.0.CO;2
- 9. Gauthier JD, Miller CR, Wilbur AE. TaqMan® MGB real-time PCR approach to quantification of Perkinsus marinus and Perkinsus spp. in oysters. J Shellfish Res 2006;25:619-24. https://doi.org/10.2983/0730-8000(2006)25[619:TMRPAT]2.0.CO;2
- 10. Rocha CSD, Sabry RC, Rocha RDS, et al. First record of Perkinsus marinus infecting Crassostrea sp. in Rio Grande do Norte, Brazil, using real-time PCR. J Invertebr Pathol 2023;198:107917. https://doi.org/10.1016/j.jip.2023.107917
- 11. Kim SH, Bathige SDNK, Kim HJ, et al. A highly sensitive and specific real-time quantitative polymerase chain reaction assay for Perkinsus marinus detection in oysters. Sci Rep 2024;14:25475. https://doi.org/10.1038/s41598-024-76822-y
- 12. Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother 2009;15:62-9. https://doi.org/10.1007/s10156-009-0669-9
- 13. Aliotta JM, Pelletier JJ, Ware JL, et al. Thermostable Bst DNA polymerase I lacks a 3′ → 5′ proofreading exonuclease activity. Genet Anal 1996;12:185-95. https://doi.org/10.1016/S1050-3862(96)80005-2
- 14. Nazina TN, Tourova TP, Poltaraus AB, et al. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int J Syst Evol Microbiol 2001;51:433-46. https://doi.org/10.1099/00207713-51-2-433
- 15. Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 2000;28:E63. https://doi.org/10.1093/nar/28.12.e63
- 16. Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes 2002;16:223-9. https://doi.org/10.1006/mcpr.2002.0415
- 17. Pumford EA, Lu J, Spaczai I, et al. Developments in integrating nucleic acid isothermal amplification and detection systems for point-of-care diagnostics. Biosens Bioelectron 2020;170:112674. https://doi.org/10.1016/j.bios.2020.112674
- 18. Biswas G, Sakai M. Loop-mediated isothermal amplification (LAMP) assays for detection and identification of aquaculture pathogens: current state and perspectives. Appl Microbiol Biotechnol 2014;98:2881-95. https://doi.org/10.1007/s00253-014-5531-z
- 19. Feng C, Wang C, Lin X, et al. Development of a loop-mediated isothermal amplification method for detection of Perkinsus spp. in mollusks. Dis Aquat Organ 2013;104:141-8. https://doi.org/10.3354/dao02591
- 20. Qu P, Wang CM, Ren WC, et al. Establishment and application of a loop-mediated isothermal amplification (LAMP) method for Perkinsus olseni detection. J Fish China 2012;36:1281-9.
- 21. Wang C, Feng CY, Wang QL, et al. Establishment of duplex loop-mediated isothermal application method for simultaneous detection of shellfish infected with Perkinsus and Bonamia. China Anim Husb Vet Med 2015;42:1935-42.
- 22. Gajamange D, Kim SH, Choi KS, Azevedo C, Park KI. Scanning electron microscopic observation of the in vitro cultured protozoan, Perkinsus olseni, isolated from the Manila clam, Ruditapes philippinarum. BMC Microbiol 2020;20:238. https://doi.org/10.1186/s12866-020-01926-0
- 23. Zhao B, Kim SH, Koh DW, et al. Molecular phylogeny, distribution, and pathogenicity of a novel thraustochytrid protist in the Manila clam, Ruditapes philippinarum, on the west and south coasts of Korea. Aquaculture 2023;575:739779. https://doi.org/10.1016/j.aquaculture.2023.739779
- 24. World Organization for Animal Health. Infection with Perkinsus marinus. In: World Organization for Animal Health. Manual of diagnostic tests for aquatic animals. The Organization; 2022.
- 25. Yang Q, Domesle KJ, Ge B. Loop-mediated isothermal amplification for Salmonella detection in food and feed: current applications and future directions. Foodborne Pathog Dis 2018;15:309-31. https://doi.org/10.1089/fpd.2018.2445
- 26. Zararsiz G, Akyildiz HY, Göksülük D, Korkmaz S, Öztürk A. Statistical learning approaches in diagnosing patients with nontraumatic acute abdomen. Turk J Electr Eng Comput Sci 2016;24:3685-97. https://doi.org/10.3906/elk-1501-181
- 27. Villalba A, Reece KS, Ordás MC, Casas SM, Figueras A. Perkinsosis in molluscs: a review. Aquat Living Resour 2004;17:411-32. https://doi.org/10.1051/alr:2004050
- 28. Ferrara M, Logrieco AF, Moretti A, Susca A. A loop-mediated isothermal amplification (LAMP) assay for rapid detection of fumonisin producing Aspergillus species. Food Microbiol 2020;90:103469. https://doi.org/10.1016/j.fm.2020.103469
- 29. Rakhmat P, Saepuloh U, Darusman HS. Development of phenol red colorimetric RT-LAMP assay in high-buffered SARS-CoV-2 sample. Hayati J Biosci 2023;30:621-31. https://doi.org/10.4308/hjb.30.4.621-631
- 30. Alhamid G, Tombuloglu H, Al-Suhaimi E. Development of loop-mediated isothermal amplification (LAMP) assays using five primers reduces the false-positive rate in COVID-19 diagnosis. Sci Rep 2023;13:5066. https://doi.org/10.1038/s41598-023-31760-z
- 31. Kreitlow A, Becker A, Ahmed MFE, et al. Combined loop-mediated isothermal amplification assays for rapid detection and one-step differentiation of Campylobacter jejuni and Campylobacter coli in meat products. Front Microbiol 2021;12:668824. https://doi.org/10.3389/fmicb.2021.668824
- 32. Tone K, Fujisaki R, Yamazaki T, Makimura K. Enhancing melting curve analysis for the discrimination of loop-mediated isothermal amplification products from four pathogenic molds: Use of inorganic pyrophosphatase and its effect in reducing the variance in melting temperature values. J Microbiol Methods 2017;132:41-5. https://doi.org/10.1016/j.mimet.2016.10.020
- 33. Hodgetts J. Rapid sample preparation and LAMP for phytoplasma detection. Methods Mol Biol 2019;1875:187-201. https://doi.org/10.1007/978-1-4939-8837-2_15
- 34. Boza JM, Erickson DC. Comparison and optimization of simple DNA extraction methods for LAMP-based point-of-care applications employing submillimeter skin biopsies. ACS Omega 2024;9:38855-63. https://doi.org/10.1021/acsomega.4c05025
- 35. Goudoudaki S, Kambouris ME, Manoussopoulou M, et al. Fast and sustainable thermo-osmotic DNA extraction protocol for trans-spectrum contingency and field use. Bio Protoc 2023;13:e4796. https://doi.org/10.21769/BioProtoc.4796
 , Seung-Hyeon Kim1
, Seung-Hyeon Kim1 , Donghyun Lee1
, Donghyun Lee1 , Hyung-Bae Jeon3
, Hyung-Bae Jeon3 , Yu Chen1,2
, Yu Chen1,2 , Kyung-Il Park1,2,*
, Kyung-Il Park1,2,*