Abstract
Recently, emerging waterborne protozoa, such as microsporidia, Cyclospora, and Cryptosporidium, have become a challenge to human health worldwide. Rapid, simple, and economical detection methods for these major waterborne protozoa in environmental and clinical samples are necessary to control infection and improve public health. In the present study, we developed a multiplex PCR test that is able to detect all these 3 major waterborne protozoa at the same time. Detection limits of the multiplex PCR method ranged from 101 to 102 oocysts or spores. The primers for microsporidia or Cryptosporidium used in this study can detect both Enterocytozoon bieneusi and Encephalitozoon intestinalis, or both Cryptosporidium hominis and Cryptosporidium parvum, respectively. Restriction enzyme digestion of PCR products with BsaBI or BsiEI makes it possible to distinguish the 2 species of microsporidia or Cryptosporidium, respectively. This simple, rapid, and cost-effective multiplex PCR method will be useful for detecting outbreaks or sporadic cases of waterborne protozoa infections.
-
Key words: Cyclospora, Cryptosporidium, microsporidia, multiplex PCR, waterborne protozoa
INTRODUCTION
Emerging waterborne protozoa, such as microsporidia,
Cyclospora, and
Cryptosporidium, have become a challenge to human health worldwide. These protozoans have several common characteristics biologically. Their major habitat is intestinal epithelial cells, and they are all intracellular parasites [
1]. In addition, they produce infectious spores that are excreted from the hosts in their stools [
2]. Although these protozoa are a concern for AIDS-infected individuals, they are gaining recognition as important infective organisms in immunocompetent individuals as well [
1,
3,
4].
Of AIDS patients with chronic diarrhea, about 50% are diagnosed as infected with microsporidia, and 10-20% are infected with
Cryptosporidium parvum [
1]. A large waterborne outbreak of
C. parvum occurred in 1993 [
5], and outbreaks of
Cyclospora that was due to consumption of imported raspberries have been reported [
6,
7]. Microsporidia has been confirmed as a waterborne protozoon based on its detection in tertiary sewage effluent, surface water, and ground water [
8]. Among the various genera in microsporidia,
Enterocytozoon bieneusi and
Encephalitozoon intestinalis are associated with gastrointestinal diseases in humans [
8].
So far, there are very limited data available pertaining to the infection status of microsporidia,
Cyclospora, and
Cryptosporidium in Korea. Development of a simple, rapid, and economical detection method for these waterborne protozoa is urgently necessary to elucidate the infection rate in a community as well as individuals. Although previously Orlandi and Lampel [
9] suggested PCR method for detection of these waterborne protozoa, their method has several disadvantages, such as demanding too much PCR reaction volume and providing no information on
Cryptosporidium hominis infection.
We developed a multiplex PCR-based method, which is simple, rapid, and cost-effective for detecting these 3 kinds of major waterborne protozoa.
MATERIALS AND METHODS
E. intestinalis spores were purchased from ATCC (Cat. No. 50506; Manassas, Virginia, USA) and maintained through in vitro culture using BS-C-1 cells (African green monkey kidney cells; Korean Cell Line Bank, Seoul, Korea) following the manufacturer's instructions. Briefly, Eagle's minimum essential medium (Sigma, St. Louis, Missouri, USA) with 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 0.1 mM non-essential amino acid, and 1.0 mM sodium pyruvate supplemented with 3% fetal bovine serum (GIBCO, Invitrogen, Carlsbad, California, USA) was used for maintenance of infection, and cells were incubated at 37℃ with 5% CO
2. Oocysts of
Cryptosporidium parvum (KKU isolate) were obtained from laboratory mice (C57BL6/J) that were infected and maintained as described [
10]. The study was approved by the Animal Care and Use Committee of Konkuk University (Seoul, Republic of Korea).
For DNA extraction from fecal specimens, QIAquick stool mini kit (QIAGEN Inc., Valencia, California, USA) was used. For
E. intestinalis and
C. parvum DNA, each number of isolated spores or oocysts from 1 × 10
4 to 1 × 10
0 were spiked into 500 µg of uninfected human stool and mixed with 10 ml distilled water (DW). After washing with DW twice by centrifugation at 100 g, pellets were resuspended in ASL lysis buffer included in the kit. The samples were incubated at 70℃ for 30 min. The procedure was executed in accordance with the manufacturer's recommendations. The extracted DNA was used as a template for PCR. DNA of
Cyclospora cayetanensis was extracted from the stool of an infected patient who was the subject of a published case study in 2003 [
11]. We cloned the 18S rRNA gene of
Cyclospora using pGEM-T easy vector (Promega, Madison, Wisconsin, USA) and used it as a template DNA after diluted it equivalent to each number of parasite.
All of the PCR primers used in this study were designed using Primer-BLAST (
http://blast.ncbi.nlm.nih.gov) and are shown in
Table 1. The first-round multiplex PCR amplification was performed in a 70-µl volume containing 5 µl of template DNA, 10 mM Tris-HCl (pH 8.3), 2 mM MgCl
2, 200 µM each of dATP, dCTP, dGTP, and dTTP, 0.7-1 µM of each primer (0.7 µM for microsporidia and 1 µM for
Cyclospora and
Cryptosporidium), and 5 U Taq DNA polymerase (Promega). The second-round multiplex PCR amplification was performed under the same conditions, except 2 µl of template DNA (product of the first-round PCR) was used in a 30-µl final volume. Both amplifications were performed using a C-1000 DNA thermal cycler (Bio-Rad, Hercules, California, USA) with initial denaturation at 94℃ for 5 min, followed by 35 cycles at 94℃ for 30 sec, 53℃ for 30 sec, and 72℃ for 90 sec and a final extension at 72℃ for 10 min. The second-round PCR cycling conditions were identical to the first-round PCR except for the annealing temperature (55℃). Amplified DNA was analyzed by electrophoresis in a 2% (w/v) agarose gel stained with ethidium bromide (0.5 µg/ml) and visualized under ImageQuant300 (GE Healthcare, Giles, UK).
An aliquot (5 µl) of the second-round PCR product of E. intestinalsis or C. parvum was used for enzyme digestion with BsaBI or BsiEI (New England BioLabs, Massachusetts, USA) at 60℃ for 2 hr. DNA fragments were analyzed by electrophoresis in a 2% (w/v) agarose gel as described above.
RESULTS
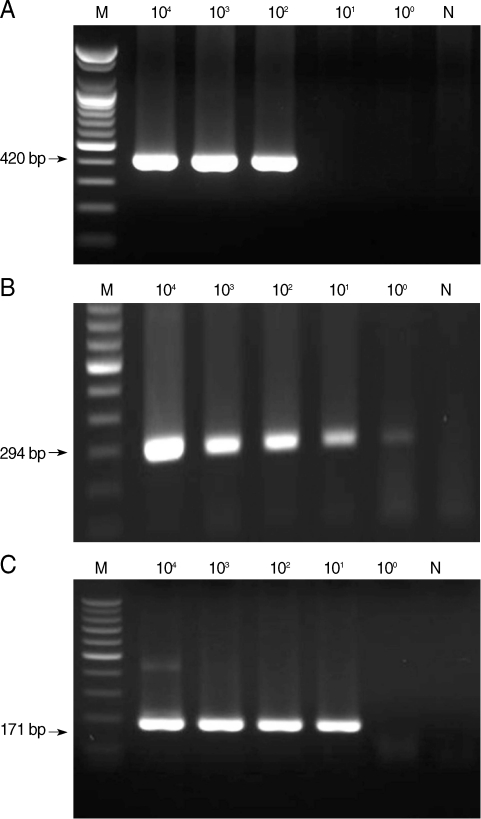
From the first-round PCR amplification, we obtained the following products with the predicted sizes: 644-657 bp, 636 bp, and 415-427 bp for microsporidia,
C. cayetanensis, and
Cryptosporidium, respectively (data not shown). From the second-round PCR, the following predicted PCR products were amplified: 410-420 bp, 294 bp, and 171-183 bp for microsporidia,
C. cayetanensis, and
Cryptosporidium, respectively (
Fig. 1). The lower detection limit of nested PCR amplification that was performed with template DNA from each individual protozoa was 10
2 spores for microsporidia, 10
0 oocysts for
Cyclospora, and 10
1 oocysts for
Cryptosporidium (
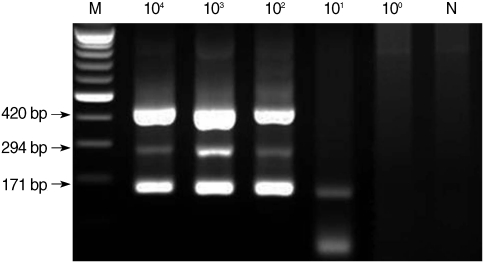
Fig. 1). The lower detection limit of multiplex PCR amplification that was performed with mixed DNA from each protozoan was 10
2 for microsporidia and
Cyclospora and 10
1 for
Cryptosporidium (
Fig. 2).
The primers for microsporidia and
Cryptosporidium used in the present study are capable of detecting both
E. bieneusi and
E. intestinalis, and
C. parvum and
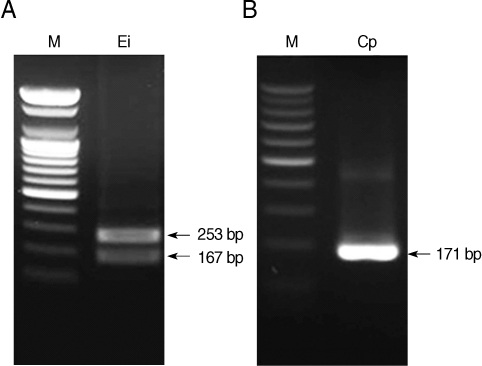
C. hominis, respectively. Restriction enzyme digestion of the resulting nested PCR products can distinguish the 2 kinds of microsporidia and
Cryptosporidium spp. Restriction enzyme digestion with BsaBI showed 2 fragmented bands of 167 and 253 bp in
E. intestinalis (
Fig. 3A), and with BsiEI showed no fragmented bands in
C. parvum (
Fig. 3B). Whereas it was confirmed with Clone Manager 6 (Sci-Ed, North Carolina, USA) that there will be no fragmented bands in
E. bieneusi with BsaBI digestion and 2 fragmented bands of 48 and 137 bp in
C. hominis with BsiEI digestion. Each primer set for microsporidia,
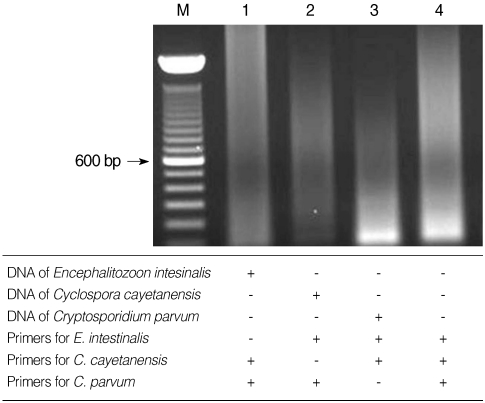
C. cayetanensis, and
Cryptosporidium used in the present study showed no cross-reactivity with each of the other parasite DNA (
Fig. 4).
DISCUSSION
There are various detection methods currently available for microsporidia,
Cyclospora, and
Cryptosporidium. These include morphological examination after specific staining, fluorescence microscopy after specific antibody labeling, and molecular diagnosis with such methods as PCR [
2,
12,
13]. Although it is inexpensive and fast, the classical staining method depends on highly trained experts for accurate diagnosis. Immunofluorescence-based antibody-labeling methods depend on the sensitivity and specificity of antibodies against parasites for the accurate diagnosis, and cross-reactivity or nonspecific labeling can be a challenge. Multiplex PCR would eliminate the need for a highly trained expert and would reduce the diagnostic time by being able to catch 3 kinds of protozoa at the same time. If well-designed multiplex PCR having a high sensitivity and specificity is available, it would be an excellent diagnostic method for these waterborne protozoa.
The primers used for
Cyclospora in this study were designed by Relman et al. [
14]. Orlandi and Lampel [
9] reported a sensitivity level for
Cyclospora that was as low as 3 oocysts using nested PCR with the same primer set, which is similar to the level of sensitivity in our study. In addition, Orlandi and Lampel [
9] described a detection sensitivity of
C. parvum and
E. intestinalis of as few as 10 oocysts from singleplex PCRs with each parasite DNA [
9]. Their detection sensitivity for
E. intestinalis is 10-fold higher than our results; however, they did not report the sensitivity of multiplex PCR using mixed DNA of each protozoan. Furthermore, agarose gel electrophoresis analysis of PCR products from both the first and second rounds of PCR is necessary to confirm the detection results with the method described by Olandi and Lampel [
9], because that method amplified the
C. parvum target only during the first round of PCR.
Generally the sensitivity of multiplex PCR is not exactly the same with individual PCR done with each parasite DNA. Multiplex PCR developed in the present study showed the same detection sensitivity with that of individual PCR in case of microsporidia and Cryptosporidium. Agarose gel electrophoresis of the second-round PCR products could provide all of the detection results, when the multiplex PCR tests reported here were applied.
Another advantage of the multiplex PCR test developed in the present study is that the 2 species of microsporidia, such as E. bieneusi and E. intestinalis, and the 2 species of Cryptosporidium, such as C. parvum and C. hominis, can be differentiated after PCR by restriction enzyme digestion. We did not show restriction enzyme-digested PCR products of E. bieneusi and C. hominis in the present study as we did not secure DNAs of these 2 organisms. Instead, we confirmed the results of restriction enzyme digestion of these 2 organisms after PCR using a restriction enzyme cutting software program (Clone Manager 6).
Although there is a well-established genomic database for
Cryptosporidium (
http://cryptodb.org) that was established through the efforts of many devoted and excellent scientists, there are still only limited genomic data available for
Cyclospora and microsporidia. Therefore it is quite difficult to develop specific primers for these 2 protozoa. The difficulty is especially challenging for
Cyclospora because PCR detection of the 18S rRNA gene could be confused with that of
Eimeria spp., which show 98% similarity with the 18S rRNA gene sequence of
Cyclospora. Thus sequence analysis after multiplex PCR should be performed for the accurate differential diagnosis of
Cyclospora and
Eimeria spp.
In conclusion, the multiplex PCR test developed in the present study can detect microsporidia, Cyclospora, and Cryptosporidium, all of which cause severe waterborne diarrheal disease. The test is very simple and rapid and offers high sensitivity and specificity. This test could help improve detection of diarrheal outbreaks or sporadic diarrheal disease that is due to infection with these major waterborne protozoa which have been considered as unknown etiology for the difficulty in detection previously.
ACKNOWLEDGEMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (RF) funded by the Ministry of Education, Science, and Technology (2010-0017321).
References
- 1. Goodgame RW. Understanding intestinal spore-forming protozoa: cryptosporidia, microsporidia, isospora, and cyclospora. Ann Intern Med 1996;124:429-441.
- 2. O'Donoghue PJ. Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol 1995;25:139-195.
- 3. Sandfort J, Hannemann A, Gelderblom H, Stark K, Owen RL, Ruf B. Enterocytozoon bieneusi infection in an immunocompetent patient who had acute diarrhea and who was not infected with the human immunodeficiency virus. Clin Infect Dis 1994;19:514-516.
- 4. Lee JK, Song HJ, Yu JR. Prevalence of diarrhea caused by Cryptosporidium parvum in non-HIV patients in Jeollanam-do, Korea. Korean J Parasitol 2005;43:111-114.
- 5. Mac Kenzie WR, Hoxie NJ, Proctor ME, Gradus MS, Blair KA, Peterson DE, Kazmierczak JJ, Addiss DG, Fox KR, Rose JB, et al. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N Engl J Med 1994;331:161-167.
- 6. Herwaldt BL, Ackers ML. An outbreak in 1996 of cyclosporiasis associated with imported raspberries. The Cyclospora Working Group. N Engl J Med 1997;336:1548-1556.
- 7. Ortega YR, Roxas CR, Gilman RH, Miller NJ, Cabrera L, Taquiri C, Sterling CR. Isolation of Cryptosporidium parvum and Cyclospora cayetanensis from vegetables collected in markets of an endemic region in Peru. Am J Trop Med Hyg 1997;57:683-686.
- 8. Dowd SE, Gerba CP, Pepper IL. Confirmation of the humanpathogenic microsporidia Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Vittaforma corneae in water. Appl Environ Microbiol 1998;64:3332-3335.
- 9. Orlandi PA, Lampel KA. Extraction-free, filter-based template preparation for rapid and sensitive PCR detection of pathogenic parasitic protozoa. J Clin Microbiol 2000;38:2271-2277.
- 10. Lee SU, Joung M, Ahn MH, Huh S, Song H, Park WY, Yu JR. CP2 gene as a useful viability marker for Cryptosporidium parvum. Parasitol Res 2008;102:381-387.
- 11. Yu JR, Sohn WM. A case of human cyclosporiasis causing traveler's diarrhea after visiting Indonesia. J Korean Med Sci 2003;18:738-741.
- 12. Didier ES. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop 2005;94:61-76.
- 13. Shields JM, Olson BH. Cyclospora cayetanensis: a review of an emerging parasitic coccidian. Int J Parasitol 2003;33:371-391.
- 14. Relman DA, Schmidt TM, Gajadhar A, Sogin M, Cross J, Yoder K, Sethabutr O, Echeverria P. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J Infect Dis 1996;173:440-445.
Fig. 1Sensitivity of nested PCR primer sets for microsporidia (A), Cyclospora cayetanensis (B), and Cryptosporidium (C). M, DNA marker; N, negative control (DW).
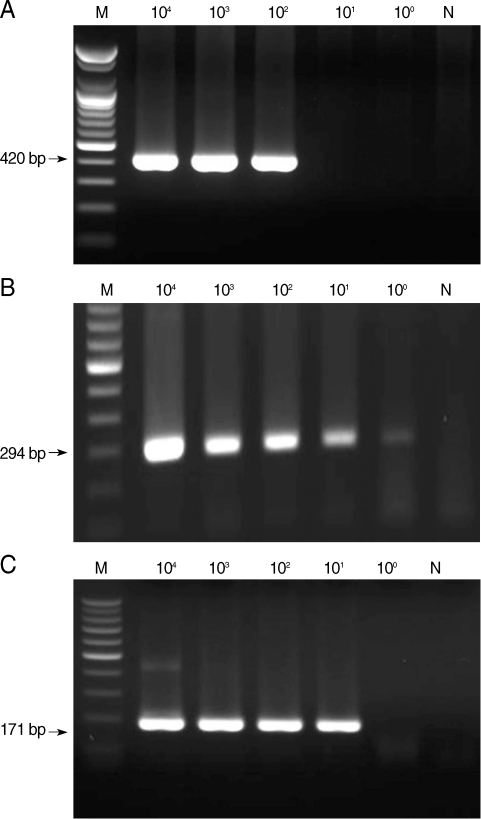
Fig. 2Sensitivity of multiplex nested PCR for microsporidia, Cyclospora cayetanensis, and Cryptosporidium. Serially diluted template DNA from each type of protozoan was mixed, and 3 kinds of primer sets for each type were included in 1 reaction tube. M, DNA marker; N, negative control (DW).
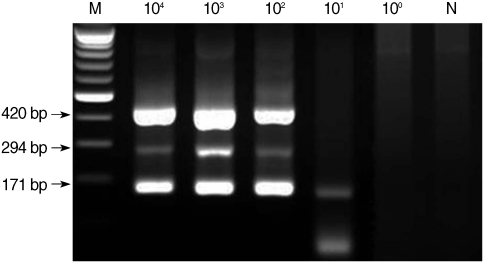
Fig. 3Restriction enzyme digestion of nested PCR products of microsporidia digested with BsaBI (A) and Cryptosporidium parvum digested with BsiEI (B). M, DNA marker; Ei, E. intestinalis; Cp, C. parvum.
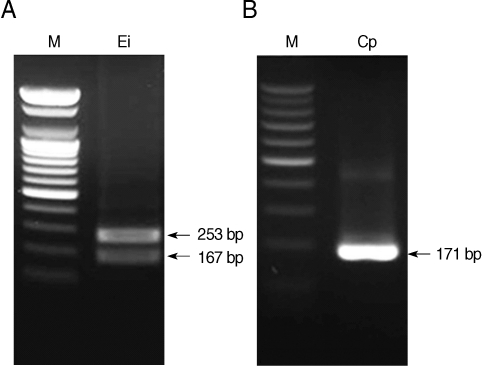
Fig. 4Specificity of multiplex nested PCR primers. No cross-reacted PCR bands were detected when the 3 primer sets were paired with DNA from the different protozoa. Lane 4, negative control (DW).
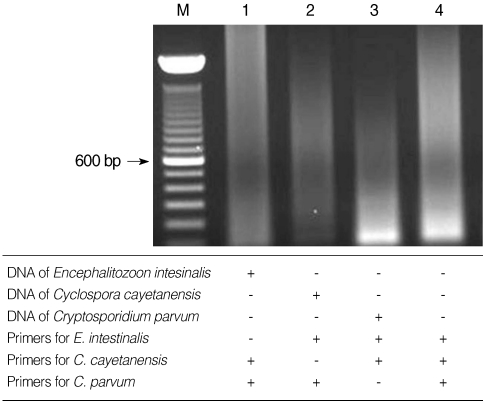
Table 1.Primer sets used for multiplex PCR
Table 1.
|
Parasite |
PCR |
Primer name |
Sequence |
PCR product size |
|
Microsporidia |
1st |
Mic A |
5′-GGAGCCTGAGAGATGGCT-3′ |
644-657 bp |
|
|
Mic E |
5′-AACGGCCATGCACCAC-3′ |
|
|
2nd |
Mic C |
5′-GGTGCCAGCAGCCGCGG-3′ |
410-420 bp |
|
|
Mic D |
5′-GCACAATCCACTCCT-3′ |
|
|
Cyclospora cayetanensis
|
1st |
CYCF1E |
5′-TACCCAATGAAAACAGTTT-3′ |
636 bp |
|
|
CYCR2B |
5′-CAGGAGAAGCCAAGGTAGG-3′ |
|
|
2nd |
CYCF3E |
5′-CCTTCCGCGCTTCGCTGCGT-3′ |
294 bp |
|
|
CYCR4B |
5′-CGTCTTCAAACCCCCTACTG-3′ |
|
|
Cryptosporidium spp. |
1st |
cp2415F |
5′-CCCACGCGAAGTTGAAGTAAC-3′ |
415-427 bp |
|
|
cp2415R |
5′-CTTAGGTTGCTTGCTTGGAGTTGG-3′ |
|
|
2nd |
cp2171F |
5′-CAACCCAGAAGTTGAGGTT-3′ |
171-183 bp |
|
|
cp2171R |
5′-CTAGTATGCTTCAGACCATGAG-3′ |
|