Abstract
Acanthamoeba infection is difficult to treat because of the resistance property of Acanthamoeba cyst against the host immune system, diverse antibiotics, and therapeutic agents. To identify encystation mediating factors of Acanthamoeba, we compared the transcription profile between cysts and trophozoites using microarray analysis. The DNA chip was composed of 12,544 genes based on expressed sequence tag (EST) from an Acanthamoeba ESTs database (DB) constructed in our laboratory, genetic information of Acanthamoeba from TBest DB, and all of Acanthamoeba related genes registered in the NCBI. Microarray analysis indicated that 701 genes showed higher expression than 2 folds in cysts than in trophozoites, and 859 genes were less expressed in cysts than in trophozoites. The results of real-time PCR analysis of randomly selected 9 genes of which expression was increased during cyst formation were coincided well with the microarray results. Eukaryotic orthologous groups (KOG) analysis showed an increment in T article (signal transduction mechanisms) and O article (posttranslational modification, protein turnover, and chaperones) whereas significant decrement of C article (energy production and conversion) during cyst formation. Especially, cystein proteinases showed high expression changes (282 folds) with significant increases in real-time PCR, suggesting a pivotal role of this proteinase in the cyst formation of Acanthamoeba. The present study provides important clues for the identification and characterization of encystation mediating factors of Acanthamoeba.
-
Key words: Acanthamoeba, encystation, microarray, cystein proteinase
INTRODUCTION
Acanthamoeba is the causative agent of granulomatous amoebic encephalitis, a fatal disease of the central nervous system, and amoebic keratitis, a painful sight-threatening disease of the eyes [
1]. The life cycle of
Acanthamoeba can be considered in 2 stages, trophozoites and cysts. Under unfavorable conditions, the trophozoite forms a cyst with high resistance to various biocides, pH, and high temperature [
2]. Encystation is one of the most common causes of treatment failure in
Acanthamoeba infection.
Recently, to understand the encystation mechanism of cyst-forming protozoa, encystation mediating factors have been investigated. Especially in
Acanthamoeba, the mechanism of the cyst-specific protein 21 during differentiation of
Acanthamoeba was reported [
3], a serine proteinase mediating encystation of
Acanthamoeba was characterized [
4], and the major role of cysteine proteases during the early phase of
Acanthamoeba encystment was suggested [
5]. The autophagy related protein 8 (Atg8) mediating autophagy formation, and autophagy related protein 3-mediated lipidation of Atg8, involved in encystation of
Acanthamoeba, were characterized [
6,
7]. To examine the gene expression profile during encystation of
Acanthamoeba castellanii, differentially expressed gene (DEG) screening was performed [
8], and an expressed sequence tag (EST) analysis and eukaryotic orthologous groups (KOG) assignment of encystation induced genes were investigated [
9,
10]. In addition, EST database of
Acanthamoeba cysts and trophozoites for comparative gene studies was constructed [
11]. These previous studies could help researchers understand the encystation mechanisms of encysting protozoa, including
Acanthamoeba. However, the exact comparison of the expression profile of cysts and trophozoites has yet to be elucidated. Further investigation to understand the encystation mechanisms requires exact data of DEGs during encystation to find the encystation mediating factors.
In this study, we report on cDNA microarray analysis containing 12,544 genes of
Acanthamoeba by comparing the transcriptome of cysts and trophozoites. Microarray analysis can monitor simultaneously gene expression levels of a large number of genes [
12]. Previously, we have reported ESTs analysis and KOG assignment of
Acanthamoeba cysts and trophozoites [
9,
10]. Based on ESTs from our
Acanthamoeba ESTs database [
11],
Acanthamoeba ESTs of TBest DB [
13], and other
Acanthamoeba related genes in the NCBI, cDNA microarray chip was constructed and differentially expressed genes between cysts and trophozoites were analyzed. This is the first report in cyst forming protozoa, including
Acanthamoeba to use the cDNA microarray technique to research encystation mediating genes.
MATERIALS AND METHODS
Amoebae cultivation
A. castellanii Castellani (ATCC #30011) was obtained from the American Type Culture Collection and was grown axenically in PYG (Peptone-Yeast-Glucose) at 25℃ [
9].
Induction of encystation and preparation of total RNA
Encystment was induced according to the procedure of Bowers and Korn [
14]. After 3 days, the cells were harvested for extraction of the total RNA. Total RNAs of trophozoites and cysts were purified using TRIzol reagent (Gibco BRL, Rockville, Maryland, USA).
The MessageAmp™ II-Biotin Enhanced (Ambion, Austin, Texas, USA) Single Round aRNA Amplification Kit is based on the RNA amplification protocol developed in the laboratory of James Eberwine [
15]. The procedure consists of reverse transcription with an oligo (dT) primer bearing a T7 promoter using ArrayScript™, a reverse transcriptase (RT) engineered to produce higher yields of first strand cDNA than wild type enzymes. ArrayScript catalyzes the synthesis of virtually full-length cDNA, which is the best way to ensure production of reproducible microarray samples. The cDNA then undergoes a second strand synthesis and clean-up to become a template for in vitro transcription in a reaction containing biotin-modified UTP and T7 RNA polymerase. To maximize biotin-labeled aRNA yield, an optimized mixture of biotin-labeled and unlabeled NTPs was supplied with kit, and Ambion's proprietary MEGAscript® in vitro transcription (IVT) technology was used to generate hundreds to thousands of antisense RNA copies of each mRNA in a sample.
Hybridization was performed with 5 µg of a labeled target sample per 1 CustomArray™ 12K microarray. Hybridizations were performed for 16 hr at 45℃ with gentle rotation. Thereafter, the arrays were washed with 6X SSPE, 0.05% Tween -20 for 5 min, 3X SSPE, 0.05% Tween -20 for 1 min, 0.5X SSPE, 0.05% Tween -20 for 1 min, and 2X PBS, 0.1% Tween -20 for 1 min. Biotin-labeled targets were hybridized for 30 min with gentle rotation. Thereafter, the arrays were washed with 2X PBS, 0.1% Tween -20 for 1 min, and 2X PBS for 1 min. The LifterSlip™ was laid at an angle onto the microarray so that it was centered over the semiconductor area. First the Imaging Solution was touched with one side of the LifterSlip™, and then slowly the slip down was lowered, taking care not to introduce air bubbles.
Microarray data acquisition
Hybridized microarrays were scanned with PMT 250-270, Pixel size 5, Focus position 130, using a GenePix 4200A microarray scanner (Axon Instruments, Union City, New Jersey, USA). After completion of scan, the resultant image was examined by the eye and proceeded for data extraction (
https://webapps.combimatrix.com).
After data extraction, background for individual samples was calculated. For the background calculation, factory built control probes with low intensities (lowest 5-30%) was used, and then their median signal intensity that can be used for subtraction was calculated. The microarray data for individual samples were normalized by a mean-shift normalization using the probes with signal value greater than zero, signal value lesser than 60,000 (saturation value), and signal value greater than the lowest 5% of each sample' signal value. A total of 7,825 probes (or 7,862 probes) were used in the final analysis. LPE-test P-value of <0.05 and Fold-change (>2, <-2) were applied to determine differentially expressed sets of genes across 3 experimental groups. We also performed complete linkage Hierarchical clustering based on Euclidean distance measure of samples using the normalized significant genes. We looked into the patterns of expressed changes for groups. We used ArrayAssist® 5.5.1 (Stratagene, La Jolla, California, USA) as statistical software.
Gene expression analysis based on real-time PCR
Nine target genes were selected for real-time PCR, and 18s rDNA was used for the reference gene (sense 5'-TCCAATTTTCTGCCACCGAA and antisense 5'-ATCATTACCCTAGTCCTCGCGC). Target gene-specific primers were defined for each transcript with the ABI primer express software (
Table 2). Total RNA was purified using Trizol reagent (Gibco BRL, Rockville, Maryland, USA), and cDNA synthesis was conducted using a RevertAid first-strand cDNA synthesis kit (Fermentas, Hanover, Maryland, USA). Real-time PCR was performed with the ABI PRISM® 7000 sequence detection system (Applied Biosystems, Foster City, California, USA), using the default thermocycler program for all genes; 10 min of preincubation at 95℃, followed by 40 cycles of 15 sec at 95℃, and 1 min at 60℃. Individual reactions were carried out in 20 µl volumes in a 96-well plate containing 1 × buffer, 3.5 mM MgCl
2, 0.2 mM deoxynucleoside triphosphates, different concentrations of sense and antisense primers (
Table 2), 0.025 U/µl enzyme, and SYBR green. All reaction mixtures were made using SYBR Premix Ex Taq (Takara, Otsu, Shiga, Japan). The experiments were performed in triplicate.
RESULTS
Features of cDNA chip and expressed cluster analysis
A cDNA microarray was constructed with 12,544 genes based on
Acanthamoeba ESTs database [
11], TBest DB [
13], and all
Acanthamoeba related genes in the NCBI, and spotted in duplicate on each microarray slide. To check the reproducibility between samples, the scatter plot comparisons of gene expression pattern between samples were performed (data not shown). Between cyst and cyst, and trophozoite and trophozoite, samples had a similarity value of 1 approximately. This showed the perfect positive correlation. However, between cysts and trophozoites, samples had a similarity value of 0.9 approximately. These identified genes appeared to be differentially expressed between cysts and trophozoites. Hierarchical clustering of the data obtained from 12,544 elements was analyzed (data not shown). Red areas indicate up-regulated transcripts and green areas indicate down-regulated transcripts in the cyst than in the trophozoite. The color saturation degree corresponded to the gene expression ratio. Gene transcripts that were up-regulated or down-regulated more than 2-folds were considered to be differentially expressed. Comparison of the signals obtained using the microarray appreciated the fold change in gene expression for 1,560 genes. Up-regulated transcripts in the cyst compared to the trophozoite were 701 genes, while down-regulated transcripts were 859 genes.
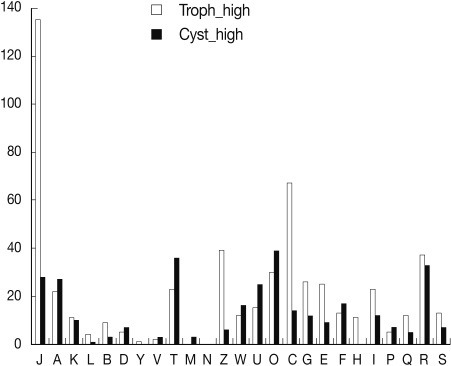
A general functional classification of 1,560 differentially expressed genes was obtained by a BLASTX similarity search against the KOG database (
Table 1). A (RNA processing and modification), P (inorganic ion transport and metabolism), D (cell cycle control, cell division, and chromosome partitioning), V (defense mechanisms), T (signal transduction mechanisms), W (extracellular structures), U (intracellular trafficking, secretion, and vesicular transport), and O (posttranslational modification, protein turnover, and chaperones) articles were also more highly populated in cyst genes than those of trophozoites. Comparison of the KOG analysis between microarray results and previous ESTs analysis [
9,
10] revealed higher percentages of cyst genes related to D, T, and O articles than those of trophozoites. Especially, KOG analysis of microarray showed a higher percentage of cyst genes to T and O articles, and a lower percentage of cyst genes related to J (translation, ribosomal structure, and biogenesis) and C (energy production and conversion) articles than those of trophozoites (
Fig. 1). Microarray analysis also suggests that signal molecules and protein modification related proteins are important in encystation of
Acanthamoeba.
To validate the microarray results, quantitative analysis of 9 genes was performed by real-time PCR (
Table 2). To correct differences in RNA quality and quantity among the samples, data were normalized using the 18s rDNA. All of 9 genes (
Table 2) were highly expressed during encystation. These expression levels were correlated with the microarray analysis.
Microarray results indicated that 701 genes were highly expressed more than 2-folds during encystation of
Acanthamoeba.
Table 3 shows the list of genes highly expressed more than 10-folds in the cyst than in the trophozoite. As many as 66 genes were highly expressed more than 10-folds in cysts. Especially, cysteine proteinases showed significantly high expression changes, 282 folds in cysts. Real-time PCR also showed high expression levels of cysteine proteinases. On the other hand, among these genes, 22 genes were not identified and 42 genes were not annotated (#N/A). These unidentified or non-annotated genes are needed to be investigated more about functional roles in encystation of
Acanthamoeba.
DISCUSSION
This comparison of transcriptome between cysts and trophozoites in
A. castellanii using cDNA microarray demonstrated that 701 genes were highly expressed more than 2-folds in cysts, and 859 genes were expressed lower in cysts than in trophozoites. Among them, cysteine proteinases showed high expression changes (
Table 3), and real-time PCR confirmed their high expression levels during encystation (
Table 2). Leitsch et al. [
5] reported that the protease inhibitors, E64d [(2S,3S)-trans-epoxysuccinyl-L-leucylamido-3-methylbutane ethyl ester], inhibited the onset of encystment of
A. castellanii, and described the major role for cysteine proteases during the early phase of encystment [
5]. Previously identified cyst specific ESTs [
10], such as a cysteine proteinase (NC0115/282-fold increase), ubiquitin (ACL00000192/11-fold increase), subtilisin-like serine proteinase (AC-C0457/5.5-fold increase), calreticulin (AH-0894/2-fold increase), proteophosphoglycan (NC0392/31-fold increase), and cyst specific protein 21 (AC-T0782/5.2-fold increase) were confirmed by this cDNA microarray analysis.
Newly identified various cyst specific genes in this study, such as serine/threonine protein kinase (NC1001/4.6-fold increase), translation elongation factor (NC0950/4.5-fold increase), tyrosine kinase like protein (ACL00008738/4.4-fold increase), Ras-related small GTP binding protein (ACL00004031/3.9-fold increase), calcium transporting ATPase (ACL00008443/2.1-fold increase), and others were stored on the '
Acanthamoeba ESTs database' [
11]. The list of the fold changes in gene expression for 1,560 genes and their expression levels were stored in an additional file 1 (data not shown). We compared these microarray results with previous reported ESTs analysis data [
9,
10]. These results commonly indicated that the exhibited high expression changes in cyst genes were significantly enriched for the signal transduction mechanisms (T article) and posttranslational modification, protein turnover, and chaperones (O article). This analysis reveals that signal transduction mechanism is essential for the formation of cysts, and various proteinases required for unnecessary protein degradation or posttranslational modifications. Cysteine proteinases that showed high expression changes in this study may be involved in protein degradation correlated with autophagic procession during encystation.
This is the first report on transcriptional profile comparison between cysts and trophozoites of
Acanthamoeba using cDNA microarray. A comparison of the Giardia lamblia trophozoite and cyst transcriptome using microarray has been reported with higher expression of 215 genes in the cyst [
16]. However, our
Acanthamoeba microarray analysis showed 701 transcriptomes that could provide information of various encystation mediating genes. Real-time PCR results of randomly selected genes (
Table 2) also corroborated microarray data and clearly showed that this microarray data was reliable. This observation of expression differences between cysts and trophozoites could provide many interesting genes related to encystation of
Acanthamoeba. Furthermore, the microchip produced in this study may provide clues to widen the study of excystation mediating factors and genes related to virulence or other transcriptome analysis of cyst-forming protozoa, including
Acanthamoeba.
ACKNOWLEDGMENTS
This work was supported by the Korea Research Foundation Grant funded by the Korea Government (MOEHRD, Basic Research Promotion Fund) (2010-0003606) and Brain Korea 21 Project in 2010.
References
- 1. Marciano-Cabral F, Cabral G. Acanthamoeba spp. as agents of disease in humans. Clin Microbiol Rev 2003;16:273-307.
- 2. Coulon C, Collignon A, McDonnell G, Thomas V. Resistance of Acanthamoeba cysts to disinfection treatments used in health care settings. J Clin Microbiol 2010;48:2689-2697.
- 3. Chen L, Orfeo T, Gilmartin G, Bateman E. Mechanism of cyst specific protein 21 mRNA induction during Acanthamoeba differentiation. Biochim Biophys Acta 2004;1691:23-31.
- 4. Moon EK, Chung DI, Hong YC, Kong HH. Characterization of a serine proteinase mediating encystation of Acanthamoeba. Eukaryot Cell 2008;7:1513-1517.
- 5. Leitsch D, Köhsler M, Marchetti-Deschmann M, Deutsch A, Allmaier G, Duchêne M, Walochnik J. Major role for cysteine proteases during the early phase of Acanthamoeba castellanii encystment. Eukaryot Cell 2010;9:611-618.
- 6. Moon EK, Chung DI, Hong YC, Kong HH. Autophagy protein 8 mediating autophagosome in encysting Acanthamoeba. Mol Biochem Parasitol 2009;168:43-48.
- 7. Moon EK, Chung DI, Hong Y, Kong HH. Atg3-mediated lipidation of Atg8 is involved in encystation of Acanthamoeba. Korean J Parasitol 2011;49:103-108.
- 8. Moon EK, Chung DI, Hong YC, Kong HH. Differentially expressed genes of Acanthamoeba castellanii during encystation. Korean J Parasitol 2007;45:283-285.
- 9. Moon EK, Chung DI, Hong YC, Ahn TI, Kong HH. Acanthamoeba castellanii: gene profile of encystation by ESTs analysis and KOG assignment. Exp Parasitol 2008;119:111-116.
- 10. Moon EK, Chung DI, Hong Y, Kong HH. Expression levels of encystation mediating factors in fresh strain of Acanthamoeba castellanii cyst ESTs. Exp Parasitol 2011;127:811-816.
- 11. Moon EK, Kim JO, Xuan YH, Yun YS, Kang SW, Lee YS, Ahn TI, Hong YC, Chung DI, Kong HH. Construction of EST database for comparative gene studies of Acanthamoeba. Korean J Parasitol 2009;47:103-107.
- 12. Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995;270:467-470.
- 13. O'Brien EA, Koski LB, Zhang Y, Yang L, Wang E, Gray MW, Burger G, Lang BF. TBestDB: A taxonomically broad database of expressed sequence tags (ESTs). Nucleic Acids Res 2007;35:D445-D451.
- 14. Bowers B, Korn ED. The fine structure of Acanthamoeba castellanii (Neff strain). II. Encystment. J Cell Biol 1969;41:786-805.
- 15. Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A 1990;87:1663-1667.
- 16. Faghiri Z, Widmer G. A comparison of the Giardia lamblia trophozoite and cyst transcriptome using microarrays. BMC Microbiol 2011;11:91.
Fig. 1Comparison of gene expression by microarray and KOG analysis. Cyst up-regulated genes showed higher percentages in A, D, T, W, U, O, and F articles. In J, C, and Z (cytoskeleton) articles, cyst genes were down-regulated.

Table 1.KOG analysis of cyst and trophozoite genes up-regulated more than 2 folds
Table 1.
|
KOG category |
|
No. of genes (%)
|
|
Cyst-high |
Troph-high |
|
Information storage and processing |
|
|
|
|
J |
Translation, ribosomal structure and biogenesis |
28 (3.99) |
135 (15.72) |
|
A |
RNA processing and modification |
27 (3.85) |
22 (2.56) |
|
K |
Transcription |
10 (1.43) |
11 (1.28) |
|
L |
Replication, recombination and repair |
1 (0.14) |
4 (0.47) |
|
B |
Chromatin structure and dynamics |
3 (0.43) |
9 (1.05) |
|
Metabolism |
|
|
|
|
C |
Energy production and conversion |
14 (2.00) |
67 (7.80) |
|
G |
Carbohydrate transport and metabolism |
12 (1.71) |
26 (3.03) |
|
E |
Amino acid transport and metabolism |
9 (1.28) |
25 (2.91) |
|
F |
Nucleotide transport and metabolism |
17 (1.40) |
13 (1.51) |
|
H |
Coenzyme transport and metabolism |
0 (0.00) |
11 (1.28) |
|
I |
Lipid transport and metabolism |
12 (1.71) |
23 (2.68) |
|
P |
Inorganic ion transport and metabolism |
7 (1.00) |
5 (0.28) |
|
Q |
Secondary metabolites biosynthesis, transport and metabolism |
5 (0.71) |
12 (1.40) |
|
Cellular process and signaling |
|
|
|
|
D |
Cell cycle control, cell division, chromosome partitioning |
7 (1.00) |
5 (0.58) |
|
Y |
Nuclear structure |
0 (0.00) |
1 (0.12) |
|
V |
Defense mechanisms |
3 (0.43) |
2 (0.23) |
|
T |
Signal transduction mechanisms |
36 (5.14) |
23 (2.68) |
|
M |
Cell wall/membrane/envelope biogenesis |
3 (0.43) |
0 (0.00) |
|
N |
Cell motility |
0 (0.00) |
0 (0.00) |
|
Z |
Cytoskeleton |
6 (0.86) |
39 (4.54) |
|
W |
Extracellular structures |
16 (2.28) |
12 (1.40) |
|
U |
Intracellular trafficking, secretion, and vesicular transport |
25 (3.57) |
15 (1.75) |
|
O |
Posttranslational modification, protein turnover, chaperones |
39 (5.56) |
30 (3.49) |
|
Poorly characterized |
|
|
|
|
R |
General function prediction only |
33 (4.71) |
37 (4.31) |
|
S |
Function unknown |
7 (1.00) |
13 (1.51) |
|
Not classified |
|
381 (54.35) |
319 (37.14) |
|
Total |
|
701 (100.00) |
859 (100.00) |
Table 2.Real-time PCR analysis of selected genes based on the microarray results
Table 2.
|
ID |
Probe comment |
Fold difference
|
Primer sequence |
|
Microarray |
Real-time PCR |
|
NC0115-T3 |
Cysteine proteinase; ACCP2 |
282.38 |
1,384 |
F: 5´-AAC AGC ACG CTC GTT TCC CTC T-3´ |
|
|
|
|
R: 5´-GTA GTT GGC CTC CGT CAT GAG CTT-3´ |
|
ACL00008706 |
Catalase related cluster |
56.30 |
11.0 |
F: 5´-ATC GCA CAC ACA CCA CAC TTT C-3´ |
|
|
|
|
R: 5´-CAA CGG AGC ACA AGA GAA CAA A-3´ |
|
NC0392-T3 |
Proteophosphoglycan ppg4 |
31.21 |
137.19 |
F: 5´-TCA TCA TCA CCT CGC TAC ACC A-3´ |
|
|
|
|
R: 5´-TAG TTG CAG GTG CGT TCG AGA-3´ |
|
NC0903-T3 |
Arginase II |
9.27 |
20.25 |
F: 5´-CTC AAG GAG CTA GAC GGT GGA A-3´ |
|
|
|
|
R: 5´-TAG GTG TCG AGA AGG CCA GAG A-3´ |
|
AC-T0782-T3 |
Cyst specific protein CSP21 |
5.19 |
24.95 |
F: 5´-GAG AAG TAC GGC AAC GTG GAG A-3´ |
|
|
|
|
R: 5´-GCA TAG AGC AGG CGT TCA ACA-3´ |
|
ACL00008536 |
Dipeptidyl peptidase IV cluster |
4.64 |
4.06 |
F: 5´-ACT GGC GAC ACA CCG TCT ATC T-3´ |
|
|
|
|
R: 5´-TGC GCG AGG AAT CAA CAA A-3´ |
|
NC0224 |
Protein serine/threonine kinase |
3.85 |
8.17 |
F: 5´-CCA CAT CAA CCA AAT CAC CGA-3´ |
|
|
|
|
R: 5´-CGC ATA ACC GAA CAT CAT CCA-3´ |
|
ACL00001979 |
Ubiquitin |
3.69 |
13.27 |
F: 5´-ACA CAC AAG CCA GCA ATG CA-3´ |
|
|
|
|
R: 5´-AAG TGG TGG AGG AAA CAC GCT-3´ |
|
AC-C0063 |
Heat shock protein |
2.8 |
3.41 |
F: 5´-GAC CGA CGG CAA TAA ACT CGT-3´ |
|
|
|
|
R: 5´-TCG CTG CAC CAA CAG ATT CTC-3´ |
Table 3.The list of genes that showed high expression changes more than 10 fold in cysts
Table 3.
|
ID |
Probe comment |
KOG |
Fold (Cyst/troph) |
|
NC0115-T3 |
Cysteine proteinase; ACCP2 |
O |
282.38 |
|
NC0961-T3 |
Cysteine proteinase; ACCP2 |
O |
273.54 |
|
AC-T0581-T3 |
Similar to elaC homology |
R |
80.82 |
|
ACE00000784 |
- |
- |
73.39 |
|
ACL00003940 |
Herpes_BLLF1 multi-domain protein |
- |
60.94 |
|
ACL00008706 |
Catalase related cluster |
AR |
56.30 |
|
NC0692-T3 |
Extracellular endoglucanase |
WV |
54.94 |
|
NC0680-T3 |
- |
- |
53.32 |
|
ACL00002157 |
- |
- |
48.23 |
|
NC0642-T3 |
- |
- |
41.34 |
|
NC0757-T3 |
- |
- |
38.91 |
|
ACL00003760 |
- |
- |
36.86 |
|
ACL00009010 |
- |
- |
36.09 |
|
ACL00004491 |
- |
- |
33.26 |
|
NC0678-T3 |
- |
- |
31.73 |
|
NC0457-T3 |
- |
- |
31.72 |
|
NC0392-T3 |
Proteophosphoglycan ppg4 |
- |
31.21 |
|
AC-C0617-T3 |
SJCHGC01975 protein |
O |
29.79 |
|
ACL00008637 |
Novel protein related cluster |
U |
27.80 |
|
NC0271-T3 |
Salivary gland secretion 1 CG3047-PA |
- |
27.04 |
|
NC0289-T3 |
- |
- |
26.25 |
|
NC0423-T3 |
Proteophosphoglycan 5 |
- |
23.32 |
|
ACL00004246 |
- |
- |
23.31 |
|
NC0956-T3 |
Similar to serine/arginine repetitive matrix 1 |
- |
22.54 |
|
NC0802-T3 |
Proteophosphoglycan 5 |
WV |
21.72 |
|
ACL00002714 |
Dynamin B precursor related cluster |
UR |
21.23 |
|
NC0276-T3 |
Similar to conserved hypothetical protein |
T |
20.90 |
|
ACE00001346 |
Similar to molecular chaperone (HSP90 family) |
O |
20.34 |
|
NC0283-T3 |
Proteophosphoglycan ppg4 |
- |
19.68 |
|
ACL00008639 |
Herpes_gp2 multi-domain protein |
- |
19.22 |
|
NC0994-T3 |
Proteophosphoglycan 5 |
- |
19.13 |
|
NC0807-T3 |
Antifreeze glycoprotein |
- |
18.43 |
|
NC0791-T3 |
Hypothetical protein Kpol_1072p54 |
DO |
17.66 |
|
ACL00002613 |
- |
- |
17.13 |
|
NC1140-T3 |
Proteophosphoglycan ppg4 |
- |
16.94 |
|
NC0582-T3 |
- |
- |
16.92 |
|
ACL00002232 |
- |
- |
16.50 |
|
ACL00002108 |
Transcription factor TFIIF |
K |
15.49 |
|
NC0747-T3 |
Pseudogene |
- |
15.27 |
|
NC0325-T3 |
Proteophosphoglycan ppg4 |
- |
15.07 |
|
NC1103-T3 |
Proteophosphoglycan ppg4 |
- |
14.95 |
|
AC-T0529-T3 |
- |
- |
14.77 |
|
ACL00002483 |
- |
- |
14.58 |
|
AC-T0879-T3 |
Iron-sulfur cluster assembly fam... |
CU |
14.54 |
|
NC0896-T3 |
Proteophosphoglycan ppg4 |
- |
14.53 |
|
ACL00001119 |
trEST Trithorax protein related cluster |
R |
13.83 |
|
NC0987-T3 |
Palmitoyl-protein thioesterase 3 |
IO |
13.77 |
|
NC0591-T3 |
Similar to scavenger receptor class F, member 2, partial |
WV |
13.70 |
|
NC0780-T3 |
Hemerythrin HHE cation binding domain-containing protein |
- |
13.63 |
|
ACL00008833 |
Extensin_2 multi-domain protein |
T |
12.98 |
|
ACL00008542 |
- |
- |
12.47 |
|
ACL00009103 |
- |
- |
12.34 |
|
ACL00009220 |
- |
- |
11.76 |
|
NC0721-T3 |
LigA |
- |
11.57 |
|
NC0626-T3 |
Proteophosphoglycan 5 |
A |
11.57 |
|
ACL00000192 |
Ubiquitin |
OR |
11.03 |
|
NC1049-T3 |
Antifreeze glycoprotein |
- |
10.89 |
|
ACL00008353 |
Tymo_45kd_70kd multi-domain protein |
WV |
10.73 |
|
NC0528-T3 |
BTB/POZ domain-containing protein |
- |
10.62 |
|
ACL00002099 |
Putative 5-3 exoribonuclease related cluster |
LA |
10.47 |
|
ACL00008536 |
Dipeptidyl peptidase IV related cluster |
O |
10.46 |
|
ACL00003598 |
trEST Herpes_UL56 multi-domain protein |
- |
10.35 |
|
ACL00004676 |
- |
- |
10.27 |
|
AC-T0408-T3 |
Histone H3 |
B |
10.16 |
|
NC0574-T3 |
Hypothetical protein MGG_00576 |
- |
10.10 |
|
ACL00000775 |
Unknown (eubacterial similarity) |
- |
10.05 |