Abstract
Blastocystis sp. is a common zoonotic intestinal protozoa which has been classified into 17 subtypes (STs). A cross-sectional study was conducted to determine the prevalence and subtype distribution of Blastocystis in villagers living on the Thai-Myanmar border, where the risk of parasitic infection is high. A total of 207 stool samples were collected and DNA was extracted. PCR and sequencing using primers targeting small-subunit ribosomal RNA (SSU rRNA) gene were performed. The prevalence of Blastocystis infection was 37.2% (77/207). ST3 (19.8%; 41/207) was the predominant subtype, followed by ST1 (11.6%; 24/207), ST2 (5.3%; 11/207), and ST4 (0.5%; 1/207). A phylogenetic tree was reconstructed using the maximum likelihood (ML) method based on the Hasegawa-Kishino-Yano + G + I model. The percentage of bootstrapped trees in which the associated taxa clustered together was relatively high. Some sequences of Blastocystis positive samples (TK18, 39, 46, 71, and 90) were closely related to animals (pig and cattle) indicating zoonotic risks. Therefore, proper health education in parasitic prevention for the villagers should be promoted to improve their personal hygiene. Further longitudinal studies are required to monitor the prevalence of parasitic infections after providing health education and to investigate Blastocystis ST in animals living in these villages.
-
Key words: Blastocystis, zoonotic risk, Thai-Myanmar border
INTRODUCTION
Blastocystis is an intestinal protozoa found in humans and animals, as a zoonotic intestinal protozoa [
1]. It is one of the most common protozoa identified in human stool samples [
2]. The fecal-oral route of transmission is considered to be the mode. Several countries, especially developing countries, show a high infection rate of
Blastocystis [
3]. Based on gene analysis of small-subunit ribosomal RNA (
SSU rRNA), at least 17 subtypes (STs) have been identified in humans, non-human primates, mammals, and avian hosts [
4]. ST1-ST9 were found in humans [
5,
6], with ST3 being the most common subtype [
7,
8]. However,
Blastocystis ST seen in humans were also identified in animals. This suggests that animals may act as reservoirs for
Blastocystis and may be linked to zoonotic transmission. Therefore, subtyping of this organism is important to identify potential sources and routes of transmission.
Diagnosis of
Blastocystis is usually based on direct fecal examination in the laboratory. However, this method shows low sensitivity [
9] and requires an experienced and skilled microscopist. Culture methods are also available for detecting
Blastocystis infections. It is a gold standard and more sensitive than direct examination for detection of
Blastocystis parasites in stool samples [
10,
11]. However, this method is time-consuming and not routinely performed in many laboratories. PCR, using
SSU rRNA genes, provides more sensitive and rapid means for
Blastocystis detection and subtyping [
12]. It is becoming more widely used in epidemiological studies for the detection of intestinal parasites in clinical samples.
Along the border of Thailand, local people are at risk of parasitic infections because of poor quality of life and personal hygiene. Furthermore, they have a high chance of close contact with animals due to environmental and cultural conditions, which may cause zoonotic diseases in these areas. In this study, we aimed to investigate the prevalence of Blastocystis infection and the subtype distribution of Blastocystis in villagers living on the Thai-Myanmar border.
MATERIALS AND METHODS
Collection of human stool samples
A cross-sectional study was conducted at Mae La sub-district, Tha Song Yang district (Thai-Myanmar border), Tak Province, northern Thailand (
Fig. 1). This area is mountainous and difficult to visit or travel through. Therefore, villagers rarely see doctors or public health personnel. Moreover, community socio-economic status and hygiene are quite low. Physical examinations were conducted by a Mobile Clinic from the Hospital for Tropical Diseases in health services during November 2013. Total of 207 human stool samples were collected. The participants were aged from 20 to 70 years. All participants did not complain any gastrointestinal symptoms such as abdominal pain or diarrhea. However, some participants complained myalgia (muscle pain) because of their work in agriculture. The specific instructions for collecting and avoiding contamination of stool samples were clearly explained to all participants. The stool samples were kept in cool conditions during transportation, and then preserved at -20˚C until DNA extraction. The study protocol was approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand (MUTM 2014-046-01).
DNA was extracted directly from all fresh stool samples using PSP spin kit (STRATEC Inc., Berlin, Germany), following manufacturer’s instructions. Briefly, the stool samples were lysed with lysis buffer under high temperatures (95˚C) and the InviAdsorb matrix was used for eliminating undissolved particles (including PCR inhibitors) in the stool samples, followed by proteinase K enzyme to destroy some proteins and increase the purity of DNA. DNA samples were stored at -20˚C until use.
PCR amplification
The presence and characterization of
Blastocystis STs was carried out using nested PCR on the DNA from collected stool samples. The primer sets and conditions were used according to previously published articles, i.e., external primer set: RD3: GGG ATC CTG ATC CTT CCG CAG GTT CAC CTA C, RD5: GGA AGC TTA TCT GGT TGA TCC TGC CAG TA [
13], internal primer set: forward: GGA GGT AGT GAC AAT AAA TC, reverse: ACT AGG AAT TCC TCG TTC ATG [
3] and specific for the
SSU rRNA gene. The PCR products had a length of about 1,100 base pairs (bp) and were detected using 1.5% agarose gel electrophoresis. The DNA in the gel was stained with ethidium bromide and visualized under a transilluminator. PCR amplification for each sample was repeated 3 times.
All samples which were tested positive for the
Blastocystis SSU rRNA gene were sequenced using an ABI 3730 XL sequencer, fluorescent dye-terminator sequencing by Bio Basic Canada Inc. (Ontario, Canada). DNA sequences were read and edited by Bioedit. DNA sequences were compared with published sequences of the
SSU rRNA gene in 17 STs of
Blastocystis on available GenBank Database through Blast search (
http://blast.ncbi.nlm.nih.gov/Blast.cgi), as shown in
Table 1. Multiple alignments were performed by Clustal W [
14]. Phylogenetic reconstructions and calculations of genetic divergence (intra- and interspecific divergence) were performed using MEGE 5.2 [
15]. Phylogenetic trees were reconstructed using the maximum likelihood (ML) method based on Hasegawa-Kishino-Yano+G (Gamma distribution)+I (Invariant) model (best-fit substitution model). Bootstrap values were calculated by the analysis of 1,000 replicates from the ML analysis.
Proteromonas lacertae (U37108) closely related to
Blastocystis was used as an outgroup.
Descriptive analysis of percentages was used to explore the positive stool samples and subtype distributions of Blastocystis.
RESULTS
Prevalence of Blastocystis by nested PCR
The prevalence of
Blastocystis infection in this study was 37.2 % (77/207) by nested PCR method.
Fig. 2 shows the positive PCR results of
Blastocystis. The PCR products were shown to be about 1,100 base pairs (bp) long.
The subtyping of
Blastocystis was identified by direct sequencing method. ST1-ST4 were found; ST3 (19.8%; 41/207) was the predominant subtype, followed by ST1 (11.6%; 24/207), ST2 (5.3%; 11/207), and ST4 (0.5%; 1/207), respectively (
Table 2). Other STs were not detected.
The
SSU rRNA genes were analyzed for polymorphisms of the DNA sequence, which indicated 139 parsimony informative sites, 46 singleton variable sites, and 185 variable sites from a total of 1,039 sites. Intraspecific divergence and interspecific divergence ranged from 0.01-2.1% and 3.5-10.8%, respectively. ML tree of
Blastocystis inferred from
SSU rRNA gene sequences was shown in
Fig. 3. The ML method is based on the Hasegawa-Kishino-Yano+G+I model. The percentage of bootstrapped trees in which the associated taxa clustered together was relatively high. From the ML tree, the
SSU rRNA gene sequences of
Blastocystis positive samples compared with 17 reference sequences from available GenBank database showed that TK27, 58, 62, and 90 were classified in ST1, TK6 in ST2, TK18, 39, 46, 71, and 100 in ST3, and TK75 in ST4. Some sequences of
Blastocystis positive samples (TK18, 39, 46, 71, and 90) were closely related to animals (pig and cattle).
DISCUSSION
In this study, the prevalence of
Blastocystis infection was 37.2%, a relatively high prevalence. One explanation may be the study area, where poor sanitation and low socio-economic status are common. The prevalence of
Blastocystis infection varies from country to country as well as within the same country. However, it has been reported that a higher prevalence occurs in developing countries than developed countries [
3,
16,
17]. There are several risk factors for
Blastocystis infection such as personal and community hygiene, geographic area, culture, and lifestyle of a population. In Thailand, where parasitic infections are still common,
Blastocystis prevalence has been reported as high as 45% [
18-
20].
Blastocystis is an intestinal protozoan parasite commonly found in humans and a range of animals such as pigs, rodents, chickens, and monkeys. The majority of
Blastocystis infections are transmitted by the fecal-oral route. The transmission can occur from humans to humans, humans to animals or animals to humans.
To date, the genetic diversity of this parasite has been investigated by
SSU rRNA gene analysis, showing at least 17 STs [
4]. ST1-ST9 have been reported in humans; ST1-ST4 have been shown to be the most common subtypes.
Blastocystis has been isolated from several kinds of animals (zoonotic STs) belonging to the same subtypes seen in humans. This suggests that animals might act as reservoir hosts for this parasite and facilitate zoonotic transmission. In this study, subtype 3 was the predominant subtype followed by subtypes 1, 2, and 4. The results of subtype distribution are similar to previous studies conducted in many countries such as Singapore, Italy, Bangladesh, and Egypt [
3,
6,
21,
22]. Surprisingly, several studies in Thailand reported that ST1 was the common subtype [
23-
25]. The explanation lies in the difference in PCR protocols between the studies, study areas, and study population, leading to different results of subtype distribution. Previous studies revealed that primers of Bȍhm-Gloning design did not amplify all
Blastocystis subtypes, especially subtype 3 [
26].
Several methods can be used to detect intestinal parasites in clinical samples. Microscopic examination is one of the choices for detecting Blastocystis infection. However, this method shows low sensitivity and specificity, leading to misdiagnosis or underestimation of the true prevalence of Blastocystis. Moreover, this parasite shows polymorphic forms and variable size in stool samples, making it difficult to diagnose. The culture method can increase sensitivity and specificity, but it is a labour-intensive and time-consuming method. PCR is an alternative method to detect Blastocystis and many other intestinal parasites in epidemiological studies, offering an increase in sensitivity and specificity as well as rapid diagnosis. This method can also characterize the subtypes of Blastocystis and reveal the source of infection.
In this study, we used primer sets of nested PCR according to Clark [
13] (primary PCR), and Wong et al. [
3] (secondary PCR), respectively. The PCR product of the
SSU rRNA gene was about 1,100 bp. The majority of the primer target contains highly variable regions that allow the construction of a phylogenetic tree. This primer was chosen as if the PCR product size is too short; it is unreliable to perform phylogenetic analysis. When phylogenetic trees were reconstructed based on Hasegawa-Kishino-Yano+G+I model, the results showed that samples TK18, 39, 46, and 71 (ST3) and 90 (ST1) may be related to zoonotic transmission, as shown in
Fig. 3. Most villagers sampled work in agriculture. Their traditional culture and lifestyle means they are close to other humans, animals (i.e. pigs, cattle, chickens, and rodents), and the forest, which raises their risk of infection. Many did not identify the dangers associated with contact with infected animals (pathogenic parasites). Their livestock were regularly kept under or near the houses at night. This increases the chance of parasitic infections due to frequent animal contact, allowing the assumption that zoonotic transmission is more common in rural areas than urban areas. Thathaisong et al. [
23] reported that zoonotic transmission might occur in the home for girls in Thailand, as those girls who did not wash their hands after making contact with pets had a higher risk of infection. Local rhesus monkeys were the possible source of
Blastocystis (ST2) found in children in Kathmandu, Nepal [
27].
In conclusion, the zoonotic risks of Blastocystis in villagers are considered in this study. The prevalence of Blastocystis infection was still relatively high. Therefore, the villagers should be encouraged to improve their personal hygiene, as well as having access to proper health education on parasite prevention. Further longitudinal studies will be required to monitor the prevalence of parasitic infections and impacts of possible health education, and to investigate Blastocystis STs in animals living in the villages.
Notes
-
We declare that we have no conflict of interest related to this work.
We would like to thank all the participants and staff of the Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand for their help in collecting stool samples. Thanks to Mr. Paul Adams and Mr. Tim Cole for editing the English language of the manuscript.
Fig. 1.Mae La sub-district, Tha Song Yang district (Thai-Myanmar border), Tak Province, Northern Thailand.
Fig. 2.Gel electrophoresis of PCR amplification. Lane 1, 100 bp DNA marker; Lane 2, negative control; Lane 3, positive control with Blastocystis; and Lanes 4-6, positive samples with Blastocystis.
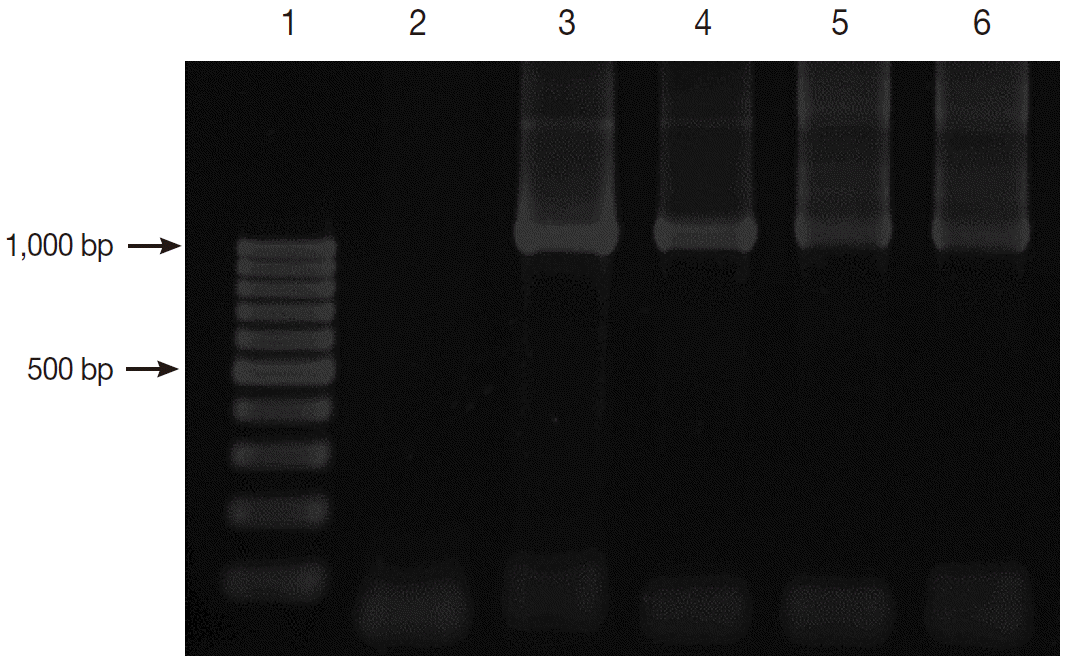
Fig. 3.Maximum-likelihood phylogenetic analysis among SSU rDNA sequences of Blastocystis. Bold letters are sequences identified in this study
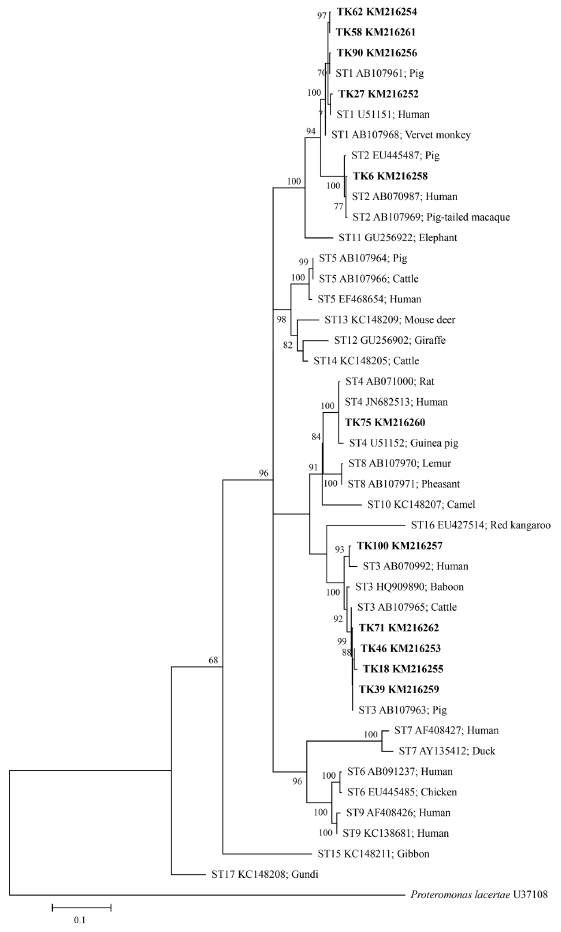
Table 1.Reference sequences of Blastocystis sp. for phylogenetic analysis in this study
Table 1.
|
Subtype |
Host |
Accession number |
Location |
|
1 |
Pig |
AB107961 |
Japan [28] |
|
Human |
U51151 |
USA [29] |
|
Vervet monkey |
AB107968 |
Japan [28] |
|
2 |
Pig |
EU445487 |
Philippines [30] |
|
Human |
AB070987 |
Japan [31] |
|
Pig-tailed macaque |
AB107969 |
Japan [28] |
|
3 |
Human |
AB070992 |
Japan [31] |
|
Baboon |
HQ909890 |
Denmark [32] |
|
Cattle |
AB107965 |
Japan [28] |
|
Pig |
AB107963 |
Japan [28] |
|
4 |
Rat |
AB071000 |
Japan [31] |
|
Human |
JN682513 |
Denmark [33] |
|
Guinea pig |
U51152 |
USA [29] |
|
5 |
Pig |
AB107964 |
Japan [28] |
|
Cattle |
AB107966 |
Japan [28] |
|
Human |
EF468654 |
China [34] |
|
6 |
Human |
AB091237 |
Japan [31] |
|
Chicken |
EU445485 |
Philippines [30] |
|
7 |
Human |
AF408427 |
Japan [31] |
|
Duck |
AY135412 |
France [35] |
|
8 |
Lemur |
AB107970 |
Japan [28] |
|
Pheasant |
AB107971 |
Japan [28] |
|
9 |
Human |
AF408426 |
Japan [22] |
|
Human |
KC138681 |
Denmark [36] |
|
10 |
Camel |
KC148207 |
UK [4] |
|
11 |
Elephant |
GU256922 |
Australia [26] |
|
12 |
Giraffe |
GU256902 |
Australia [26] |
|
13 |
Mouse deer |
KC148209 |
UK [4] |
|
14 |
Cattle |
KC148205 |
UK [4] |
|
15 |
Gibbon |
KC148211 |
UK [4] |
|
16 |
Red kangaroo |
EU427514 |
Japan [37] |
|
17 |
Gundi |
KC148208 |
UK [4] |
Table 2.Subtype distribution of Blastocystis in this study
Table 2.
|
Subtype |
Number of positive sample (%) |
|
1 |
24/207 (11.6) |
|
2 |
11/207 (5.3) |
|
3 |
41/207 (19.8) |
|
4 |
1/207 (0.5) |
|
Total |
77/207 (37.2) |
References
- 1. Tan KS. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin Microbiol Rev 2008;21:639-665.
- 2. Yakoob J, Jafri W, Beg MA, Abbas Z, Naz S, Islam M, Khan R. Irritable bowel syndrome: is it associated with genotypes of Blastocystis hominis. Parasitol Res 2010;106:1033-1038.
- 3. Wong KH, Ng GC, Lin RT, Yoshikawa H, Taylor MB, Tan KS. Predominance of subtype 3 among Blastocystis isolates from a major hospital in Singapore. Parasitol Res 2008;102:663-670.
- 4. Alfellani MA, Taner-Mulla D, Jacob AS, Imeede CA, Yoshikawa H, Stensvold CR, Clark CG. Genetic diversity of Blastocystis in livestock and zoo animals. Protist 2013;164:497-509.
- 5. Rene BA, Stensvold CR, Badsberg JH, Nielsen HV. Subtype analysis of Blastocystis isolates from Blastocystis cyst excreting patients. Am J Trop Med Hyg 2009;80:588-592.
- 6. Souppart L, Moussa H, Cian A, Sanciu G, Poirier P, El Alaoui H, Delbac F, Boorom K, Delhaes L, Dei-Cas E, Viscogliosi E. Subtype analysis of Blastocystis isolates from symptomatic patients in Egypt. Parasitol Res 2010;106:505-511.
- 7. Ozyurt M, Kurt O, Mølbak K, Nielsen HV, Haznedaroglu T, Stensvold CR. Molecular epidemiology of Blastocystis infections in Turkey. Parasitol Int 2008;57:300-306.
- 8. Roberts T, Stark D, Harkness J, Ellis J. Subtype distribution of Blastocystis isolates identified in a Sydney population and pathogenic potential of Blastocystis. Eur J Clin Microbiol Infect Dis 2013;32:335-343.
- 9. Stensvold CR, Arendrup MC, Jespersgaard C, Mølbak K, Nielsen HV. Detecting Blastocystis using parasitologic and DNA-based methods: a comparative study. Diagn Microbiol Infect Dis 2007;59:303-307.
- 10. Leelayoova S, Taamasri P, Rangsin R, Naaglor T, Thathaisong U, Mungthin M. In-vitro cultivation: a sensitive method for detecting of Blastocystis hominis. Ann Trop Med Parasitol 2002;96:803-807.
- 11. Termmathurapoj S, Leelayoova S, Aimpun P, Thathaisong U, Nimmanon T, Taamasri P, Mungthin M. The usefulness of short-term in vitro cultivation for the detection and molecular study of Blastocystis hominis in stool specimens. Parasitol Res 2004;93:445-447.
- 12. Roberts T, Barratt J, Harkness J, Ellis J, Stark D. Comparison of microscopy, culture, and conventional polymerase chain reaction for detection of Blastocystis sp. in clinical stool samples. Am J Trop Med Hyg 2011;84:308-312.
- 13. Clark CG. Extensive genetic diversity in Blastocystis hominis. Mol Biochem Parasitol 1997;87:79-83.
- 14. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 2007;23:2947-2948.
- 15. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28:2731-2739.
- 16. Hirata T, Nakamura H, Kinjo N, Hokama A, Kinjo F, Yamane N, Fujita J. Prevalence of Blastocystis hominis and Strongyloides stercoralis infection in Okinawa, Japan. Parasitol Res 2007;101:1717-1719.
- 17. Horiki N, Maruyama M, Fujita Y, Yonekura T, Minato S, Keneda Y. Epidemiologic survey of Blastocystis hominis infection in Japan. Am J Trop Med Hyg 1997;56:370-374.
- 18. Saksirisampant W, Nuchprayoon S, Wiwanitkit V, Yenthakam S, Ampavasiri A. Intestinal parasitic infestations among children in an orphanage in Pathum Thani Province. J Med Assoc Thai 2003;86:S263-S270.
- 19. Taamasri P, Leelayoova S, Rangsin R, Naaglor T, Ketupanya A, Mungthin M. Prevalence of Blastocystis hominis carriage in Thai army personnel based in Chonburi, Thailand. Mil Med 2002;167:643-646.
- 20. Mungthin M, Suwannasaeng R, Naaglor T, Areekul W, Leelayoova S. Asymptomatic intestinal microsporidiosis in Thai orphans and child-care workers. Trans R Soc Trop Med Hyg 2001;95:304-306.
- 21. Meloni D, Sanciu G, Poirier P, El Alaoui H, Chabé M, Delhaes L, Dei-Cas E, Delbac F, Luigi Fiori P, Di Cave D, Viscogliosi E. Molecular subtyping of Blastocystis sp. isolates from symptomatic patients in Italy. Parasitol Res 2011;109:613-619.
- 22. Yoshikawa H, Wu Z, Kimata I, Iseki M, Karim I, Hossain AMB, Zaman V, Haque R, Takahashi Y. Polymerase chain reaction-based genotype classification among human Blastocystis hominis populations isolated from different countries. Parasitol Res 2004;92:22-29.
- 23. Thathaisong U, Siripattanapipong S, Mungthin M, Pipatsatitpong D, Tan-Ariya P, Naaglor T, Leelayoova S. Identification of Blastocystis subtype 1 variants in the Home for Girls, Bangkok, Thailand. Am J Trop Med Hyg 2013;88:352-358.
- 24. Leelayoova S, Siripattanapipong S, Thathaisong U, Naaglor T, Taamasri P, Piyaraj P, Mungthin M. Drinking water: a possible source of Blastocystis spp. subtype 1 infection in schoolchildren of a rural community in central Thailand. Am J Trop Med Hyg 2008;79:401-406.
- 25. Thathaisong U, Worapong J, Mungthin M, Tan-Ariya P, Viputtigul K, Sudatis A, Noonai A, Leelayoova S. Blastocystis isolates from a pig and a horse are closely related to Blastocystis hominis. J Clin Microbiol 2003;41:967-975.
- 26. Parkar U, Traub RJ, Vitali S, Elliot A, Levecke B, Robertson I, Geurden T, Steele J, Drake B, Thompson RC. Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet Parasitol 2010;169:8-17.
- 27. Yoshikawa H, Wu Z, Pandey K, Pandey BD, Sherchand JB, Yanagi T, Kanbara H. Molecular characterization of Blastocystis isolates from children and rhesus monkeys in Kathmandu, Nepal. Vet Parasitol 2009;160:295-300.
- 28. Abe N. Molecular and phylogenetic analysis of Blastocystis isolates from various hosts. Vet Parasitol 2004;120:235-242.
- 29. Silberman JD, Sogin ML, Leipe DD, Clark CG. Human parasite finds taxonomic home. Nature 1996;380:398.
- 30. Rivera WL. Phylogenetic analysis of Blastocystis isolates from animal and human hosts in the Philippines. Vet Parasitol 2008;156:178-182.
- 31. Arisue N, Hashimoto T, Yoshikawa H. Sequence heterogeneity of the small subunit ribosomal RNA genes among Blastocystis isolates. Parasitology 2003;126:1-9.
- 32. Stensvold CR, Clark CG. Investigation of the molecular epidemiology of Blastocystis by use of a multi-locus sequence typing system for subtype 3 (personal communication).
- 33. Stensvold CR, Alfellani M, Clark CG. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect Genet Evol 2012;12:263-273.
- 34. Yan MY, Su LS, Lai YR, Ye HJ, Liao H, Chen FG, Luo PX, Hou PZ, Lai FX. Phylogenetic analysis of Blastocystis hominis isolates in China (personal communication).
- 35. Noël C, Peyronnet C, Gerbod D, Edgcomb VP, Delgado-Viscogliosi P, Sogin ML, Capron M, Viscogliosi E, Zenner L. Phylogenetic analysis of Blastocystis isolates from different hosts based on the comparison of small-subunit rRNA gene sequences. Mol Biochem Parasitol 2003;126:119-123.
- 36. Engsbro AL, Stensvold CR. Blastocystis: to treat or not to treat...but how? Clin Infect Dis 2012;55:1431-1432.
- 37. Yoshikawa H. Phylogenetic studies of Blastocystis isolates from zoo marsupial mammals inferred from the small subunit ribosomal RNA gene sequences (personal communication).





