Abstract
The present study was performed to compare the mitochondrial genomes between 2 Spirometra tapeworms, Spirometra erinaceieuropaei and Spirometra decipiens (Cestoidea: Diphyllobothriidae), which larval stages are important etiological agents of sparganosis in humans. For each species, the full mitochondrial genome was amplified in 8 overlapping fragments using total genomic DNA purified from a single worm as the template. The mitochondrial genomes were 13,643 bp (S. erinaceieuropaei) and 13,641 bp (S. decipiens) in length and contained 36 genes; 12 protein-coding genes, 2 ribosomal RNA (rRNA, small and large subunits), and 22 transfer RNAs (tRNAs). The 12 protein-coding genes constituted 10,083 bp (S. erinaceieuropaei) and 10,086 bp (S. decipiens) of their respective mitochondrial genomes. The tRNA genes, ranging in length from 56 to 70 bp, were identified based on putative secondary structures such as the typical cloverleaf shape. A total of 23 intergenic sequences, varying from 1 to 204 bp in size, were interspersed in S. erinaceieuropaei (total, 504 bp) and S. decipiens (total, 496 bp) mtDNA. The 12 protein-coding genes of S. erinaceieuropaei and S. decipiens differed by 12.4%, whereas the overall difference in mtDNA sequence between S. erinaceieuropaei and S. decipiens was 12.9%. Thus, from the standpoint of the mitochondrial genome, S. decipiens represents a valid species that can be distinguished from S. erinaceieuropaei.
-
Key words: Spirometra erinaceieuropaei, Spirometra decipiens, mitochondrial genome
INTRODUCTION
Spirometra erinaceieuropaei and
S. decipiens are pseudophyllidean cestodes. In humans, these worms can induce sparganosis, and their medical importance is well- established. Humans can be infected by procercoid larvae in cyclops (the first intermediate host) and plerocercoid larvae in reptiles or amphibians (paratenic hosts). The genus
Spirometra was established by Mueller [
1], but this name had been used earlier by Faust et al. [
2], who proposed dividing the genus into 2 subgenera,
Diphyllobothrium and
Spirometra.
S. erinaceieuropaei was first reported by Rudolphi as
Dubium erinaceieuropaei from hedgehogs (
Erinaceus europaeus) in 1819. Diesing in 1853 recognized this larva as a sparganum and named it
Sparganum erinaceieuropaei. Molin in 1895 renamed it as
Sparganum lanceolatum, which is regarded as a synonym. These names refer to the larval stage of spirometrid species. Based on morphological characteristics described by Diesing [
2],
S. decipiens is synonymous with 4 species;
Bothriocephalus felis Creplin, 1825,
Bothriocephalus maculatus Leukart, 1848,
Dibothrium decipiens, and
Bothriocephalus decipiens Diesing, 1850. In 1929, Faust, Campbell and Kellogg reviewed the morphological characteristics of
S. erinaceieuropaei and
S. decipiens by monitoring the experimental development of adults from spargana in humans and other vertebrates.
Currently, the species classification in the genus
Spirometra is controversial. Despite the availability of reliable diagnostic morphological characters for species identification, the specieslevel taxonomy of
Spirometra spp. remains obscure. Therefore, it is necessary to devise a reliable taxonomic criterion based on morphological characteristics in combination with molecular analysis of mitochondrial DNA sequences. Mitochondrial DNA sequencing is now considered a useful molecular tool for inferences of evolutionary analysis, phylogenetic reconstruction, taxonomic identification, biogeography, population genetics, and epidemiological investigation [
3,
4]. The complete mitochondrial genomes of order Pseudophyllidea have been recently published for
Diphyllobothrium latum [
5],
Diphyllobothrium nihonkaiense [
6], and
S. erinaceieuropaei [
7].
Within the genus
Spirometra, mitochondrial DNA sequences have also been used as genetic makers for identification and characterization of genetic variation of species [
8,
9]. The mtDNA
cox1 gene sequences revealed that
S. erinaceieuropaei and
S. proliferum are distinct species [
10]. More recently, the complete mitochondrial genome of
S. erinaceieuropaei isolated from China was reported and compared to the mitochondrial genome of a Japanese isolate [
7]. However, the speciation of the parasite may need to revise careful reconsideration based on morphological analyses of
Spirometra tapeworms. The aim of this study was to provide information useful for classification of
Spirometra spp., by the mitochondrial genome of
S. erinaceieuropaei and
S. decipiens and the mitochondrial genomes with those of other cestodes.
MATERIALS AND METHODS
Worm samples
Plerocercoid larvae of
S. decipiens were collected from
Rhabdophis tigrinus tigrinus, which were donated by the Association of Wildlife Protection, and then used to infect a cat, in April 2008. Plerocercoid larvae of
S. erinaceieuropaei were collected from a 58-year-old woman, and then used to infect a dog in May 2009. Eight weeks after the infection, the cat and the dog were sacrificed, and adult worms were recovered from their intestines. Experimental animals were used following the ethical guidelines in commission of laboratory animals in Chungbuk National University (2008). The tapeworms were pressed and fixed in alcohol-formalin-acetic acid (AFA) for carmine staining. The following anatomical features of mature and gravid proglottids were observed, based on the morphological data of Faust et al. [
2]; vaginal opening, uterus, uterine pore, cirrus, genital pore, testes, and vitellaria. Gravid proglottids were preserved at -70˚C in 70% ethanol until use. A single proglottid was chopped into small pieces, and total genomic DNA was extracted using the DNeasy tissue Kit (Qiagen, Valencia, California, USA).
For each species, the full mitochondrial genome was amplified in 8 overlapping fragments using total genomic DNA purified from a single worm as the template. The overlapping fragments of
S. decipiens and
S. erinaceieuropaei mtDNA were amplified using 16 oligonucleotide primers. The primers were used to amplify the
cob-
nad2 region (Spiro-cob-F1: TTT YCA YTC TTA TTT TAC YAC TAA GAA and Spiro-nad2-R1: AYC ACA CAT ACT CCC ARC TTG GGC TAC, 3.1 kbp),
nad2-
nad1 region (Spiro-nad2-F1: TTT GGG CST YKT GTT GYR TGT GTT ATT and Spiro-nad1-R1: CCA ACC RGC ACA CAA TAA AGC ATA ACT, 1.0 kbp),
nad1-
cox1 region (Spiro-nad1-F1: TAT GCY GAG TCG GAG AGG GAG TTG GTT and Spiro-cox1-R1: AAC MCC AAT AAT CAT AGT YAC AGA ACT, 2.1 kbp),
cox1-
rrnS region (Spiro-cox1-F1: GAC TGG TAA GTT AAT TTA AAC TGT and Spiro-12S-R1: CAT CTA ACC CAA CCG TAA CAT A, 2.5 kbp),
rrnS-
nad5 region (Spiro-12S-F1: GTA TTA ATA TTT AAG CTA AGT CTA TGT GCT and Spiro-nad5-R1: AAA CRC ACC AAG CAA TTT TAT TAC AGG TRG, 3.0 kbp) and
nad5-
cob region (Spiro-nad5-F1: CYA CCT GTA ATA AAA TTG CTT GGT GYG TTT and Spiro-cob-R1: YAA ACA AAC ATG AGC TGA AAA WAC ACG AAC, 2.5 kbp). PCR and DNA sequencing was performed as described previously [
11].
Sequences were assembled and aligned using Geneious 6.1.5 program (Biomatters Co., Auckland, New Zealand). The sequenced regions were identified using BLAST searches and compared with platyhelminth sequences in the GenBank database. Protein-coding genes were identified based on similarity of inferred amino acid sequences to those of other platyhelminth mtDNAs, as well as multiple comparisons with mitochondrial gene sequences in the GenBank database. The mitochondrial genetic code of platyhelminths was used to obtain putative translational products of the mitochondrial proteincoding sequences. Two ribosomal RNA genes (12S and 16S subunits) were determined by alignments with other known rRNA genes of platyhelminths. Twenty-two putative tRNA genes were identified using the tRNAscan-SE software [
12] and anticodon sequences. The putative stem-loop structures of non-coding mitochondrial regions were inferred using the RNAdraw program [
13].
Phylogenetic analysis was conducted using PAUP 4.0 [
14]. The following mitochondrial genome sequences were used:
S. erinaceieuropaei (KJ599680; this study),
S. decipiens (KJ599679; this study),
Diphyllobothrium latum (NC_008945),
D. nihonkaiense (NC_009463),
Echinococcus granulosus (NC_008075),
E. multilocularis (NC_000928),
Hymenolepis diminuta (NC_002767),
Taenia solium (NC_004022),
T. saginata (NC_009938),
T. asiatica (NC_004826), and
T. crassiceps (NC_002647). Phylogenetic trees were constructed using the maximum likelihood (ML), maximum parsimony (MP), neighbor joining (NJ), and Bayesian inference [
15], with
Fasciola hepatica (NC_002546) and
Paragonimus westermani (NC_002354) as outgroups. Confidence values for tree branches were determined by bootstrap analyses with 1,000 replicates.
RESULTS
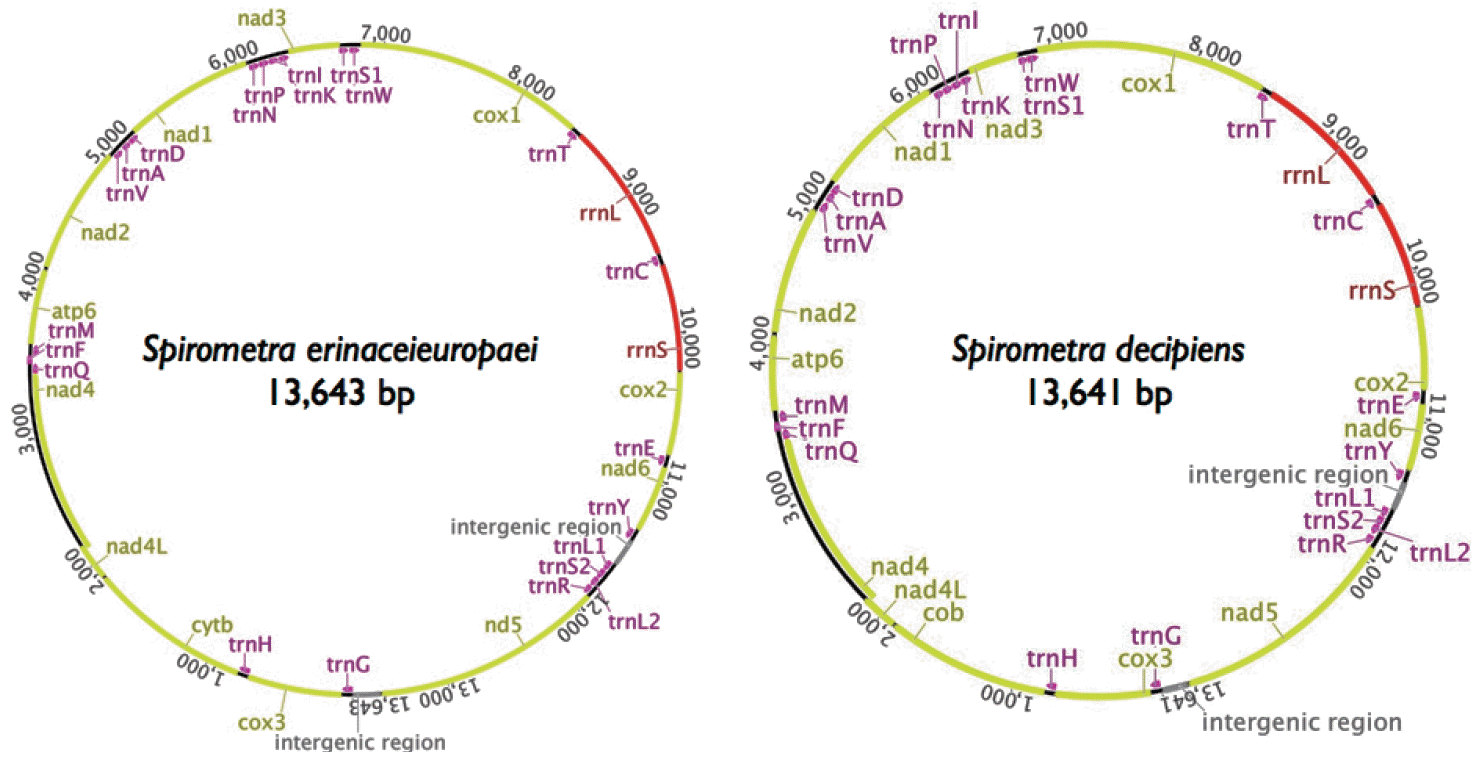
Gene content and organization
The mitochondrial genomes of
Spirometra tapeworms were 13,643 bp (
S. erinaceieuropaei) and 13,641 bp (
S. decipiens) in length. The genomes each contained 36 genes, of which 12 were protein-coding genes, 2 were ribosomal RNA (rRNAs, small and large subunit), and 22 were transfer RNAs (tRNAs) (
Fig. 1). As with other cestodes, both genomes lacked the
atp8 gene. We assumed that all the genes were transcribed on 1 strand in the same direction and arranged in the same relative positions as gene loci in known cestode mitochondrial genomes. The arrangement of mitochondrial genes in
Spirometra tapeworms was identical to that of other pseudophyllidean cestodes published to date, with the exception of
Hymenolepis diminuta, in which the order of
trnL and
trnS2 is reversed. The nucleotide compositions of the whole mitochondrial genomes are 19.8% A, 45.9% T, 23.5% G, and 10.9% C (
S. erinaceieuropaei) and 20.3% A, 46.0% T, 22.7% G, and 11.0% C (
S. decipiens). As in other cestodes, the genomes are A + T rich:
S. erinaceieuropaei, 65.7% A + T; and
S. decipiens, 66.3% A + T (
Table 1). Some genes overlap at the boundaries:
cox3/
trnH (10 bp),
nad4L/
nad4 (40 bp),
trnQ/
trnF (4 bp),
trnF/
trnM (4 bp),
nad3/
trnS1 (11 bp), and
cox1/
trnT (10 bp), respectively (
Table 2).
Approximately 65% of the mitochondrial genomes of
Spirometra tapeworms consisted of protein-encoding genes, similar to the values reported for other cestodes. The 12 protein-coding genes constituted 10,083 bp (
S. erinaceieuropaei) and 10,086 bp (
S. decipiens) of their respective mitochondrial genomes (
Table 1). All of the putative open reading frames (ORFs) of 12 protein-coding genes in both species start and end with complete codons. The ATG initiation codon was used in 11 genes (
atp6,
cob,
cox1,
cox2,
nad1,
nad2,
nad3,
nad4,
nad4L, and
nad6), whereas the GTG initiation codon was used only in the
cox3 gene. The TAG stop codon was used in 7 genes (
cox1,
cox2,
cox3,
nad2,
nad3,
nad4, and
nad4L) in
S. erinaceieuropaei and 6 genes (
cox1,
cox3,
nad2,
nad3,
nad4, and
nad4L) in
S. decipiens, whereas the TAA termination codon was used in the remaining 5 genes (
atp6,
cob,
nad1,
nad5, and
nad6) in
S. erinaceieuropaei and 6 genes (
atp6,
cob,
cox2,
nad1,
nad5, and
nad6) in
S. decipiens (
Table 2). Codon usage is shown in
Table 3. The 4 most commonly used codons were Leu (TTR and CTN; 15.6%), Phe (TTY; 12.6%), Val (GTN; 10.8%) and Ser (AGN and TCN; 9.9%) in
S. erinaceieuropaei, and Leu (TTR and CTN; 15.7%), Phe (TTY; 12.5%), Val (GTN; 10.7%), and Ser (AGN and TCN; 9.9%) in
S. decipiens.
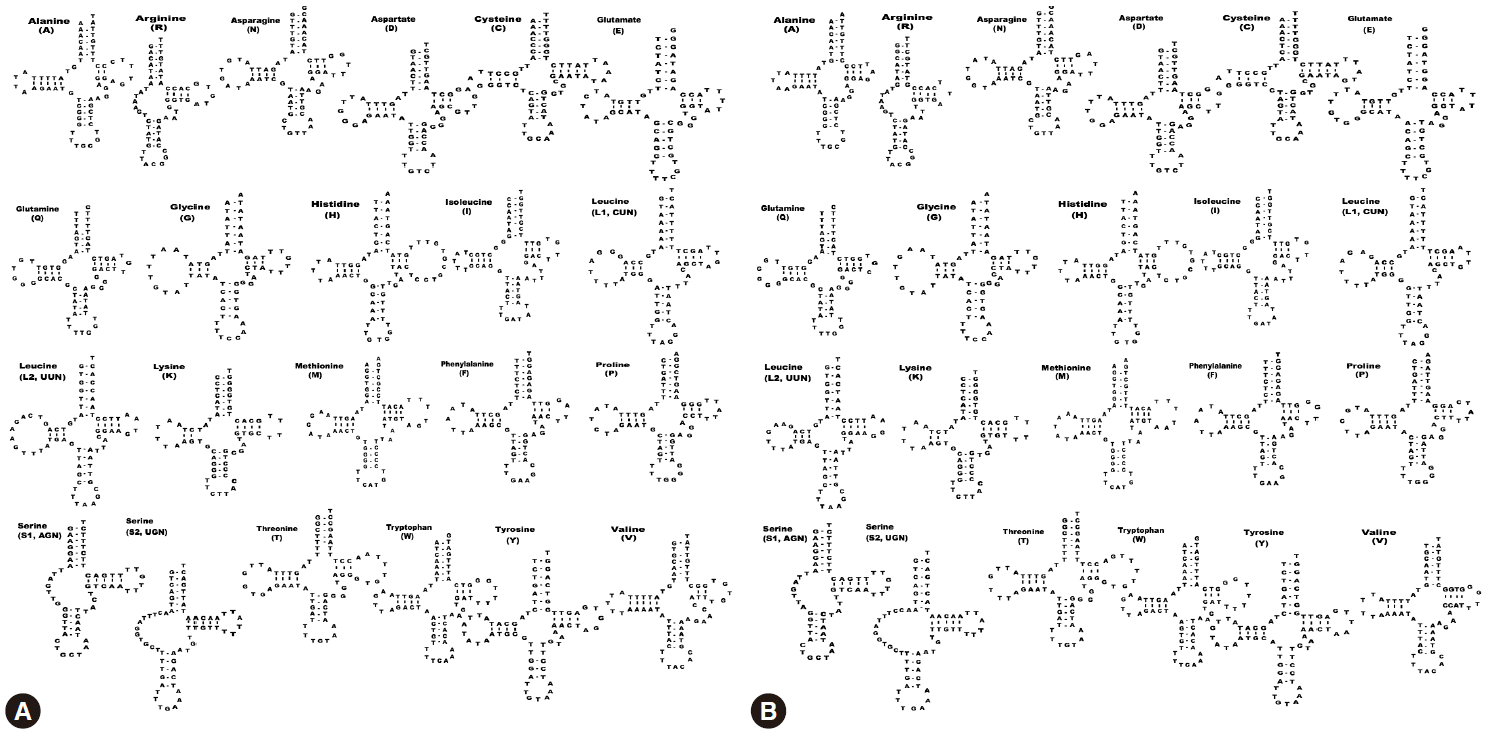
We identified a total of 22 tRNA genes in the
S. erinaceieuropaei and
S. decipiens mitochondrial genomes. The tRNA genes, ranging in length from 56 bp to 70 bp, were identified based on putative secondary structures such as the typical cloverleaf shape (
Fig. 2). The inferred secondary structure of 19 tRNA exhibited the typical cloverleaf shape, with paired DHU arms; in the remaining 3 tRNAs (
trnR,
trnS1, and
trnS2), this arm was replaced with a 7-12 nt unpaired loop. The aminoacyl acceptor arms consisted of 7 bp, and
trnA,
trnI,
trnM,
trnQ,
trnR,
trnS2,
trnT, and
trnV contained 1 or 3 non-canonical base pairs. The anticodon stems of all 22 tRNAs contained of 5 bp, as in typical stem structures. The predicted secondary structures of the 22 tRNAs in
S. erinaceieuropaei and
S. decipiens had paired TΨC arms, consisting of a 2-5 bp stem with a 3-9 bp loop. The variable loop between the anticodon and the TΨC stems consisted of 3-5 nt, except in
trnR,
trnC,
trnS1, and
trnS2 (
Fig. 2). In both species, the ribosomal RNA genes
rrnL and
rrnS were separated by
trnC. The
rrnL and
rrnS genes were 967 bp and 733 bp long, respectively, in
S. erinaceieuropaei, and 973 bp and 730 bp long in
S. decipiens. The nucleotide content of the rrn genes was 63.6% (A+T) in
S. erinaceieuropaei and 62.7% (A+T) in
S. decipiens (
Table 1).
A total of 23 intergenic sequences, varying from 1 to 204 bp in size, were interspersed in the S. erinaceieuropaei (total, 504 bp) and S. decipiens (total, 496 bp) mtDNA. Two major non-coding regions present in the mtDNA were predicted to form hairpin structures. Non-coding region 1 (NR1), between trnY and trnL1, is 201 bp (S. erinaceieuropaei) or 204 bp (S. decipiens) in length, and non-coding region 2, between nad5 and trnG was 185 bp (S. erinaceieuroapei) or 174 bp (S. decipiens) in length. The non-coding regions in these 2 species had A + T content of 70.9% (S. erinaceieuropaei) and 67.7% (S. decipiens) and contained stem-loop structures. The nucleotide contents of the NR1 and NR2 were 32.6% A, 9.3% C, 38.3% T, and 19.7% G in S. erinaceieuropaei mtDNA, while those of S. decipiens were 32.3% A, 11.9% C, 35.4% T, and 20.4% G in S. decipiens mtDNA.
Mitochondrial sequence divergence between S. erinaceieuropaei and S. decipiens
A pairwise comparison of sequence divergence of the 12 protein-coding genes and 2 ribosomal RNA genes of
S. erinaceieuropaei and
S. decipiens is shown in
Table 4. The protein-coding sequences of
S. erinaceieuroapei contained 10,083 bp and 3,361 codons, whereas those of
S. decipiens contained 10,086 bp and 3,362 codons. The 12 protein-coding genes of
S. erinaceieuropaei and
S. decipiens differed by 12.4%, whereas the full mtDNA sequences differed by 12.9%. Divergences in protein-coding genes between
S. erinaceieuropaei and
S. decipiens ranged from a low of 9.3% to a high of 13.7% (
Table 4). Amino acid sequence divergences of
cox1 (the most highly conserved gene) and
nad6 (the most variable gene) were 1.9% and 14.8%, respectively. The ribosomal RNA genes of
S. erinaceieuropaei differed by 11.2% (
rrnL) and 6.9% (
rrnS) relative to those of
S. decipiens.
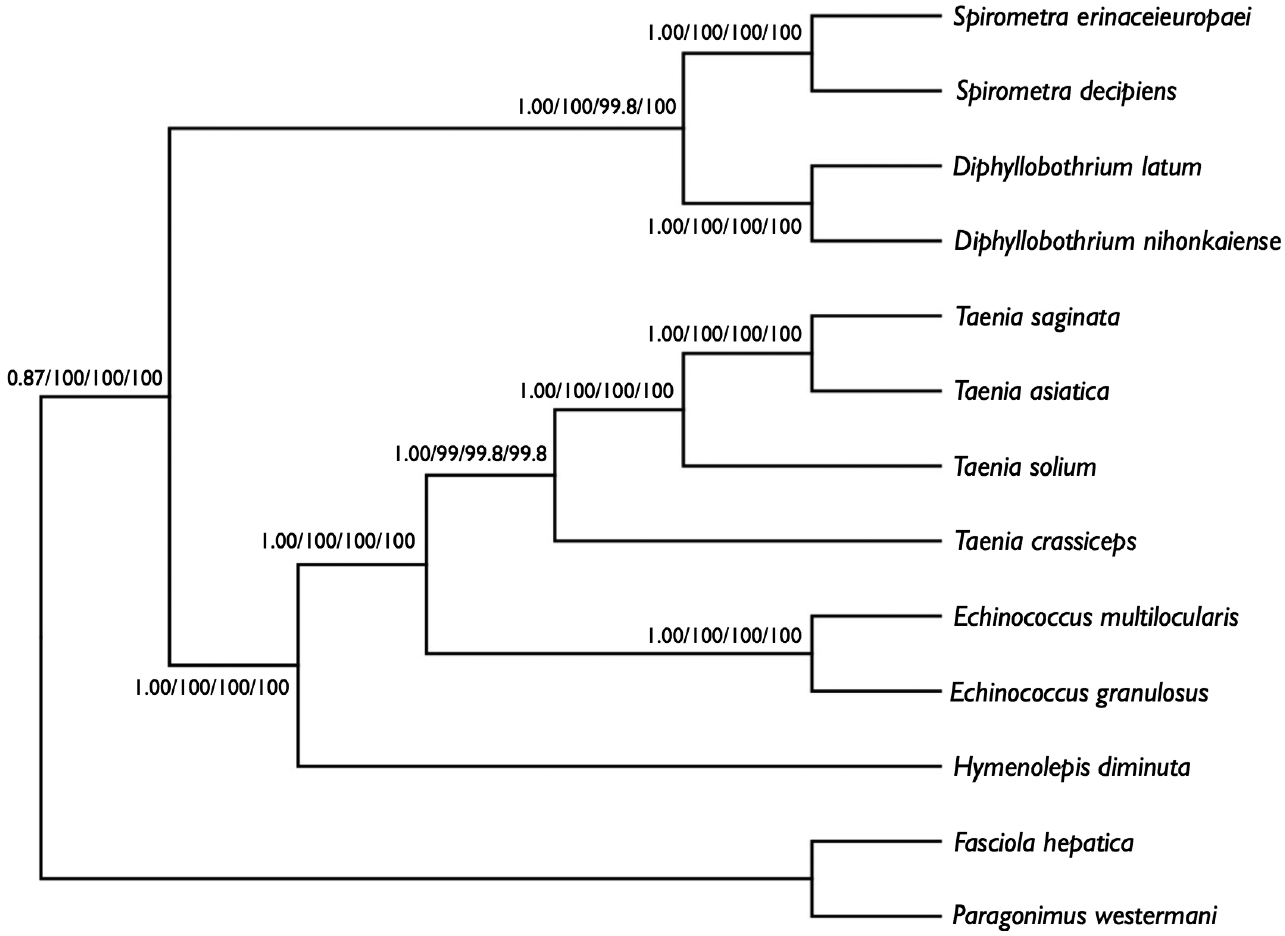
We performed phylogenetic analyses of
S. decipiens and
S. erinaceieuropaei using 4 methods (Bayesian inference, ML, NJ, and MP), based on concatenated amino acid sequences of 12 protein genes from 11 cestodes and 2 trematodes. To this end, we used an alignment set of 10,394 bp including all 12 mitochondrial protein-coding gene loci. Of the 3,141 (30.3%) homologous positions and 66.1% pairwise identity showed in the set of those mtDNA sequences from maximum likelihood analysis. A concatenated alignment set of 3,389 homologous amino acid positions and 1,425 variable sites were phylogenetically informative under MP criterion. Phylogenetic relationships among the eucestodes determined using the 4 analytic approaches exhibited identical tree topologies. In the consensus tree, order Cyclophyllidea, including family Taeniidae (
Taenia and
Echinococcus) formed a well-supported monophyletic group. Family Hymenolepididae is a sister taxon to the Taeniidae. Within the Pseudophyllidea clade,
Diphyllobothrium and
Spirometra formed a monophyletic group, and sister genera are well supported (
Fig. 3).
DISCUSSION
In the present study, we sequenced and analyzed whole mitochondrial genomes of
S. erinaceieuropaei and
S. decipiens based on the morphological analysis described in detail recently by the present authors [
16]. The full mtDNA sequences of
S. erinaceieuropaei and
S. decipiens differ by 12.9% which means
S. erinaceieuropaei and
S. decipiens are valid species that can be distinguished from each other by comparison of mitochondrial DNA sequences and morphological data as well.
Morphologically,
S. erinaceieuropaei can be clearly distinguished from
S. decipiens by its spirally coiled uterus [
2]. The uterus of
S. erinaceieuropaei consists of 5-7 complete turns, whereas that of
S. decipiens consists of 4−4½ coils. The lateral margins of the subterminal uterine coil of
S. erinaceieuropaei are parallel, while those of
S. decipiens are the broadest as subspherical in contour. In
S. erinaceieuropaei, the uterine pore lies in the midline, a small distance behind the anterior margin of the uterine terminal ball, whereas in
S. decipiens, it is a conspicuous sphincter in a ventral position under the bulge of the terminal uterine coil. The vaginal pore is a broad crescent slit behind the male genital pore in
S. erinaceieuropaei, whereas in
S. decipiens, it is crescent-shaped and elliptical. The cirrus is strongly muscular and elongated in shape in
S. decipiens, whereas in
S. erinaceieuropaei, it is smaller than in other related species [
2].
The molecular characteristics of the mitochondrial genome of S. erinaceieuropaei and S. decipiens that we identified in this study, gene arrangement, nucleotide composition, genetic code, and secondary structure of tRNA, were similar to those of other cestodes. The genetic distance between S. erinaceieuropaei and S. decipiens was determined by a percentage pairwise comparison of the nucleotide and amino acid compositions of the mitochondrial genomes. The 12 protein-coding genes of S. erinaceieuropaei and S. decipiens differed by 12.4%, whereas the sequence differences for the whole mitochondrial sequences were 12.9%. Divergences of amino acid sequences between S. erinaceieuopaei and S. decipiens ranged from a low of 1.9% (cox1) to a high of 14.8% (nad6). Therefore, these 2 parasitic organisms represent distinct species within a same genus.
These species have not been clearly identified in terms of which specifically infect humans. Conventionally,
S. erinaceieuropaei larval stages were considered to be the main reason for human infections through eating raw or undercooked snakes but actually the spargana were not seen at all naturally in the snakes. Rather,
S. decipiens has been found from snakes (unpublished data). Natural infections with
S. decipiens has been mostly observed in carnivorous animals such as cats and dogs, but no such natural infections have been observed for
S. erinaceieuropaei. Thus,
S. decipiens may only be found in dogs and cats (unpublished data). In this and the previous study [
16], adult
S. erinaceieuropaei worms were obtained by feeding dogs the larvae collected from humans or the muscle fascia of the hedgehog (
Erinaceus dealbatsu); after the initial infection, adults can be harvested in about 4 weeks. The only known second intermediate host of
S. erinaceieuropaei is the Chinese hedgehog. Therefore, in the context of human sparganosis, the infection route and relevant intermediate host of
S. erinaceieuropaei remain unclear, and there is an epidemiological discrepancy between eating habits and distributions of
Spirometra species in animals.
The information derived from the complete sequence of the
S. erinaceieuropaei and
S. decipiens mitochondrial genome will add to the available mitochondrial sequence data of parasitic cestodes, and provide a resource for comparative mitochondrial genome analyses of pseudophyllidean tapeworms. Our results show that
S. decipiens is a valid species that can be distinguished from
S. erinaceieuropaei by comparison of mitochondrial DNA sequence as well as morphological data in the previous study [
16].
Notes
-
We have no conflict of interest related to this work.
The corresponding author (Hyeong-Kyu Jeon) was supported by a Research Fellow Grant for young scientist from National Research Foundation of Korea (NRF-2012R1A1A2042993). This work was supported by a research grant from Chungbuk National University in 2014. Parasite materials used in this study were provided by the Parasite Resource Bank of Korea National Research Resource Center (2010-0003456), Republic of Korea.
Fig. 1.Schematic representation of the mitochondrial genomes of S. erinaceieuropaei and S. decipiens.
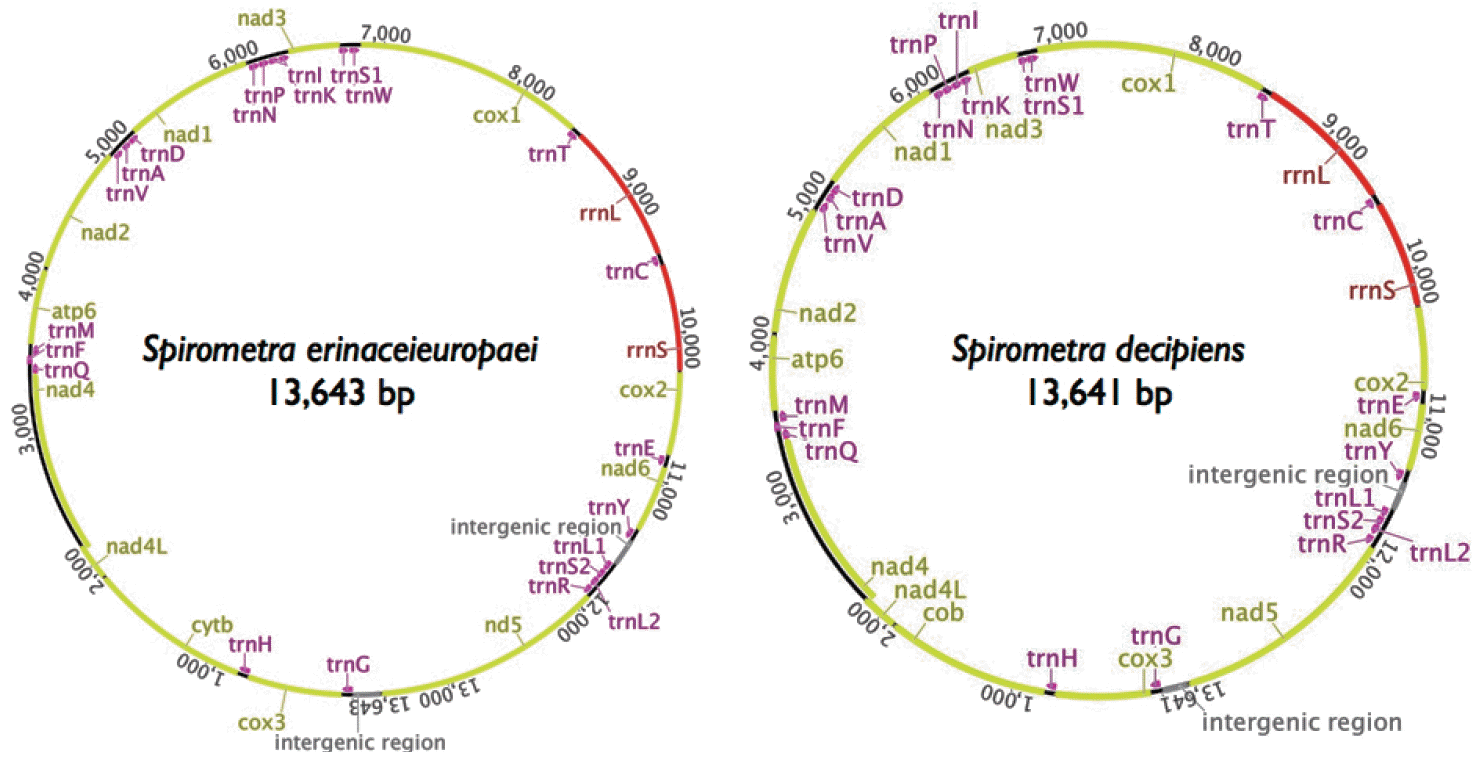
Fig. 2.Inferred secondary structures of the 22 mitochondrial tRNA from S. erinaceieuropaei (A) and S. decipiens (B).
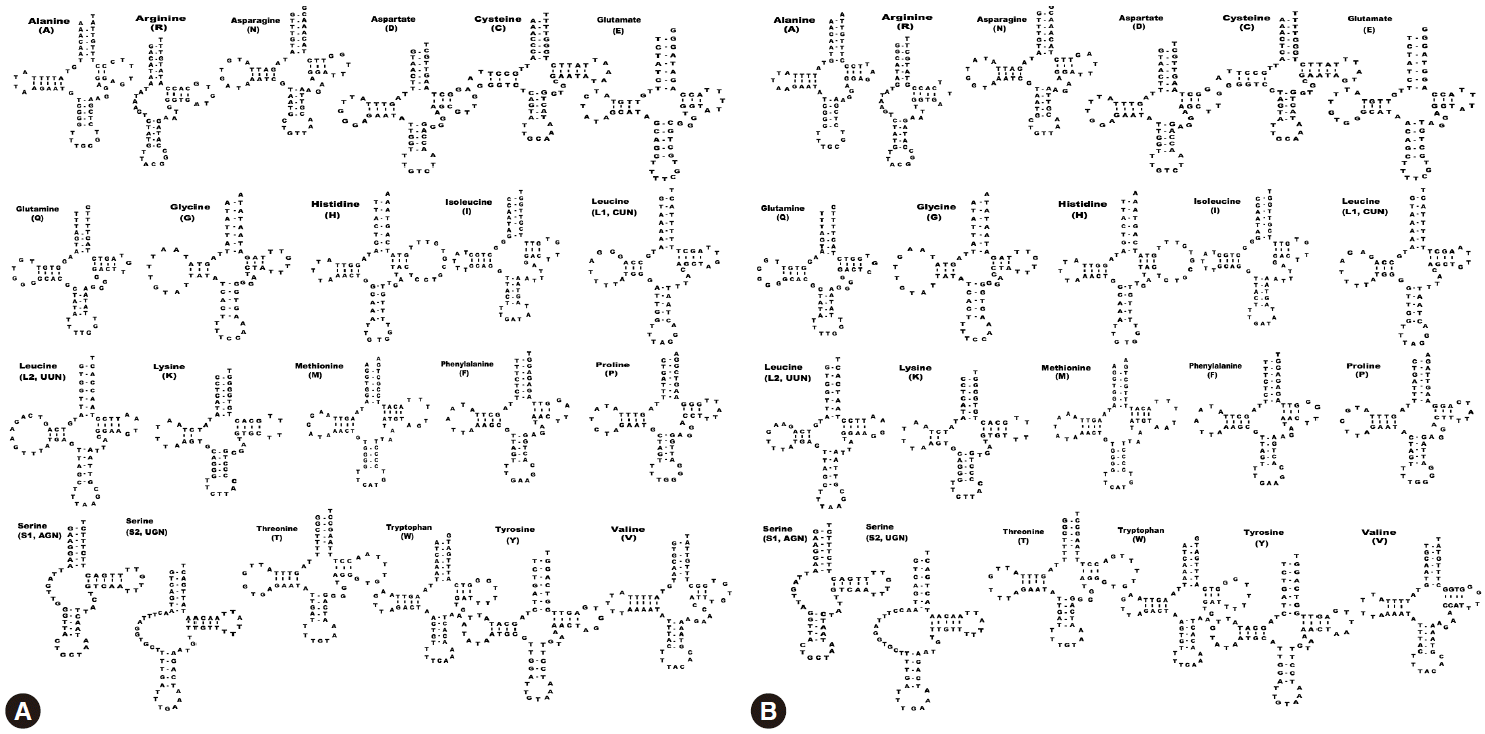
Fig. 3.Phylogenetic relationship among eucestode species based on inferred amino acid sequence data selected from 12 mitochondrial protein-coding gene loci for 13 platyhelminthes. The numbers above the branches represent bootstrap values for Bayesian inference, maximum likelihood (ML), neighbor joining (NJ), and maximum parsimony (MP), respectively.
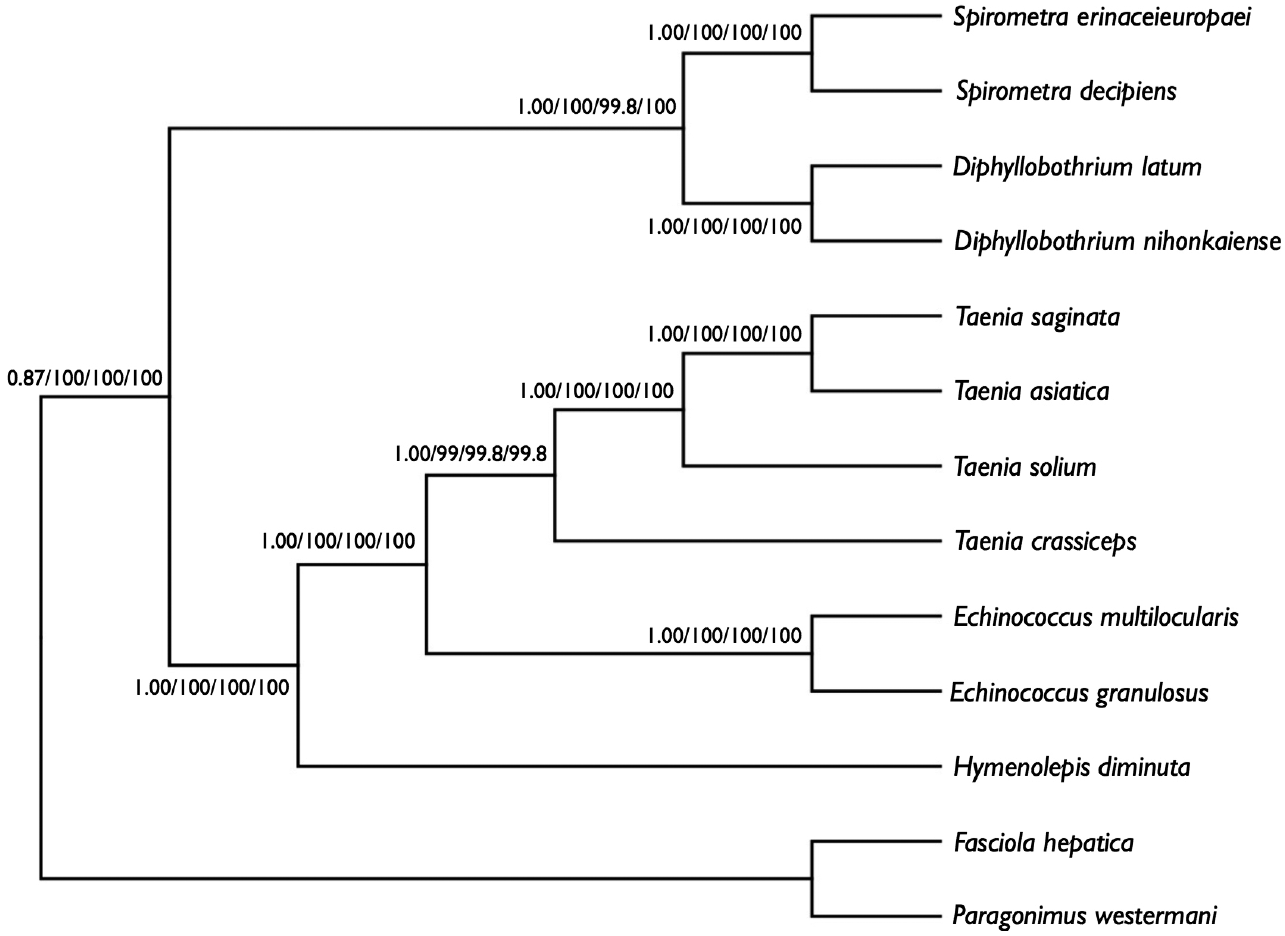
Table 1.Nucleotide compositions of the complete mitochondrial genomes, protein-coding genes, and ribosomal RNA sequences of 2 Spirometra species
Table 1.
|
Species |
Complete mtDNA sequence |
Protein-coding sequence |
rRNA sequence |
|
|
T |
C |
A |
G |
T+A |
|
T |
C |
A |
G |
T+A |
|
T |
C |
A |
G |
T+A |
|
Length (bp) |
% |
% |
% |
% |
% |
Length (bp) |
% |
% |
% |
% |
% |
Length (bp) |
% |
% |
% |
% |
% |
|
Sea
|
13,643 |
45.9 |
10.9 |
19.8 |
23.5 |
65.7 |
10,083 |
48.3 |
10.6 |
17.5 |
23.5 |
65.8 |
1,700 |
38.7 |
12.2 |
24.9 |
24.2 |
63.6 |
|
Sdb
|
13,641 |
46.0 |
11.0 |
20.3 |
22.6 |
66.3 |
10,086 |
48.6 |
10.6 |
18.3 |
22.5 |
66.9 |
1,703 |
37.6 |
12.9 |
25.1 |
24.4 |
62.7 |
Table 2.Position and characteristics of the protein-coding and non-coding sequences in the mitochondrial genomes of Spirometra erinaceieuropaei and S. decipiens
Table 2.
|
Genes |
Length of genes and sequences
|
Codon used for
|
Position in genome (5'-3') |
|
Nucleotide |
Amino acid |
Initiation |
Termination |
|
|
Sea
|
Sdb
|
Se |
Sd |
Se |
Sd |
Se |
Sd |
Se |
Sd |
|
trnG
|
67 |
67 |
|
|
|
|
|
|
1-67 |
1-67 |
|
cox3
|
651 |
651 |
216 |
216 |
GTG |
GTG |
TAG |
TAG |
71-721 |
71-721 |
|
trnH
|
70 |
69 |
|
|
|
|
|
|
712-781 |
712-780 |
|
cob
|
1,110 |
1,110 |
369 |
369 |
ATG |
ATG |
TAA |
TAA |
785-1,894 |
784-1,893 |
|
nad4L
|
261 |
261 |
86 |
86 |
ATG |
ATG |
TAG |
TAG |
1,899-2,159 |
1,898-2,158 |
|
nad4
|
1,254 |
1,254 |
417 |
417 |
ATG |
ATG |
TAG |
TAG |
2,120-3,373 |
2,119-3,372 |
|
trnQ
|
64 |
64 |
|
|
|
|
|
|
3,374-3,437 |
3,373-3,436 |
|
trnF
|
64 |
64 |
|
|
|
|
|
|
3,434-3,497 |
3,433-3,496 |
|
trnM
|
68 |
68 |
|
|
|
|
|
|
3,494-3,561 |
3,493-3,560 |
|
atp6
|
516 |
516 |
171 |
171 |
ATG |
ATG |
TAA |
TAA |
3,565-4,080 |
3,564-4,079 |
|
nad2
|
873 |
873 |
290 |
290 |
ATG |
ATG |
TAG |
TAG |
4,092-4,964 |
4,087-4,959 |
|
trnV
|
65 |
66 |
|
|
|
|
|
|
4,969-5,033 |
4,970-5,035 |
|
trnA
|
61 |
61 |
|
|
|
|
|
|
5,051-5,111 |
5,052-5,112 |
|
trnD
|
66 |
64 |
|
|
|
|
|
|
5,116-5,181 |
5,118-5,181 |
|
nad1
|
891 |
891 |
296 |
296 |
ATG |
ATG |
TAA |
TAA |
5,182-6,072 |
5,182-6,072 |
|
trnN
|
66 |
66 |
|
|
|
|
|
|
6,078-6,143 |
6,078-6,143 |
|
trnP
|
65 |
65 |
|
|
|
|
|
|
6,150-6,214 |
6,150-6,214 |
|
trnI
|
64 |
64 |
|
|
|
|
|
|
6,220-6,283 |
6,220-6,283 |
|
trnK
|
63 |
63 |
|
|
|
|
|
|
6,291-6,353 |
6,290-6,352 |
|
nad3
|
357 |
357 |
118 |
118 |
ATG |
ATG |
TAG |
TAG |
6,359-6,715 |
6,356-6,712 |
|
trnS1(AGN)
|
59 |
59 |
|
|
|
|
|
|
6,705-6,763 |
6,702-6,760 |
|
trnW
|
65 |
66 |
|
|
|
|
|
|
6,773-6,837 |
6,763-6,828 |
|
cox1
|
1,566 |
1,566 |
521 |
521 |
ATG |
ATG |
TAG |
TAG |
6,845-8,410 |
6,836-8,401 |
|
trnT
|
69 |
70 |
|
|
|
|
|
|
8,401-8,469 |
8,392-8,461 |
|
rrnL
|
967 |
973 |
|
|
|
|
|
|
8,470-9,436 |
8,462-9,434 |
|
trnC
|
65 |
65 |
|
|
|
|
|
|
9,437-9,501 |
9,435-9,499 |
|
rrnS
|
733 |
730 |
|
|
|
|
|
|
9,502-10,234 |
9,500-10,229 |
|
cox2
|
570 |
570 |
189 |
189 |
ATG |
ATG |
TAG |
TAA |
10,235-10,804 |
10,230-10,799 |
|
trnE
|
65 |
65 |
|
|
|
|
|
|
10,810-10,874 |
10,805-10,869 |
|
nad6
|
465 |
468 |
154 |
155 |
ATG |
ATG |
TAA |
TAA |
10,879-11,343 |
10,874-11,341 |
|
trnY
|
68 |
68 |
|
|
|
|
|
|
11,350-11,417 |
11,348-11,415 |
|
NR1c
|
201 |
204 |
|
|
|
|
|
|
11,418-11,618 |
11,416-11,619 |
|
trnL1(CUN)
|
67 |
67 |
|
|
|
|
|
|
11,619-11,685 |
11,620-11,686 |
|
trnS2(UGN)
|
66 |
66 |
|
|
|
|
|
|
11,688-11,753 |
11,689-11,754 |
|
trnL2(UUN)
|
65 |
65 |
|
|
|
|
|
|
11,757-11,821 |
11,759-11,823 |
|
trnR
|
56 |
57 |
|
|
|
|
|
|
11,831-11,886 |
11,839-11,895 |
|
nad5
|
1,570 |
1,569 |
522 |
522 |
ATG |
ATG |
TAA |
TAA |
11,890-13,458 |
11,899-13,467 |
|
NR2c
|
184 |
174 |
|
|
|
|
|
|
13,459-13,643 |
13,468-13,641 |
Table 3.Codon usage in the 12 protein-coding genes of the mitochondrial genomes of Spirometra species
Table 3.
|
NCc
|
AAd
|
Sea
|
Sdb
|
NC |
AA |
Se
|
Sd
|
|
No.e
|
% |
No. |
% |
No. |
% |
No. |
% |
|
TTT |
Phe |
399 |
11.9 |
386 |
11.5 |
TAT |
Tyr |
158 |
4.7 |
179 |
5.3 |
|
TTC |
Phe |
24 |
0.7 |
34 |
1.0 |
TAC |
Tyr |
42 |
1.2 |
25 |
0.7 |
|
TTA |
Leu |
187 |
5.6 |
209 |
6.2 |
TAA
|
*f
|
5 |
0.1 |
6 |
0.2 |
|
TTG |
Leu |
225 |
6.7 |
190 |
5.7 |
TAG
|
* |
7 |
0.2 |
6 |
0.2 |
|
CTT |
Leu |
58 |
1.7 |
73 |
2.2 |
CAT |
His |
44 |
1.3 |
38 |
1.1 |
|
CTC |
Leu |
8 |
0.2 |
4 |
0.1 |
CAC |
His |
10 |
0.3 |
12 |
0.4 |
|
CTA |
Leu |
19 |
0.6 |
25 |
0.7 |
CAA |
Gln |
5 |
0.1 |
4 |
0.1 |
|
CTG |
Leu |
28 |
0.8 |
26 |
0.8 |
CAG |
Gln |
18 |
0.5 |
18 |
0.5 |
|
ATT |
Ile |
142 |
4.2 |
142 |
4.2 |
AAT |
Asn |
51 |
1.5 |
54 |
1.6 |
|
ATC |
Ile |
16 |
0.5 |
14 |
0.4 |
AAC |
Asn |
10 |
0.3 |
7 |
0.2 |
|
ATA |
Ile |
64 |
1.9 |
74 |
2.2 |
AAA |
Asn |
30 |
0.9 |
36 |
1.1 |
|
ATG
|
Met |
79 |
2.4 |
80 |
2.4 |
AAG |
Lys |
47 |
1.4 |
46 |
1.4 |
|
GTT |
Val |
179 |
5.3 |
192 |
5.7 |
GAT |
Asp |
64 |
1.9 |
54 |
1.6 |
|
GTC |
Val |
22 |
0.7 |
18 |
0.5 |
GAC |
Asp |
5 |
0.1 |
14 |
0.4 |
|
GTA |
Val |
55 |
1.6 |
46 |
1.4 |
GAA |
Glu |
12 |
0.4 |
18 |
0.5 |
|
GTG
|
Val |
107 |
3.2 |
98 |
2.9 |
GAG |
Glu |
50 |
1.5 |
49 |
1.5 |
|
TCT |
Ser |
122 |
3.6 |
120 |
3.6 |
TGT |
Cys |
122 |
3.6 |
125 |
3.7 |
|
TCC |
Ser |
13 |
0.4 |
18 |
0.5 |
TGC |
Cys |
13 |
0.4 |
9 |
0.3 |
|
TCA |
Ser |
35 |
1.0 |
40 |
1.2 |
TGA |
Trp |
24 |
0.7 |
38 |
1.1 |
|
TCG |
Ser |
18 |
0.5 |
16 |
0.5 |
TGG |
Trp |
75 |
2.2 |
57 |
1.7 |
|
CCT |
Pro |
39 |
1.2 |
47 |
1.4 |
CGT |
Arg |
44 |
1.3 |
47 |
1.4 |
|
CCC |
Pro |
25 |
0.7 |
19 |
0.6 |
CGC |
Arg |
2 |
< 0.1 |
2 |
< 0.1 |
|
CCA |
Pro |
11 |
0.3 |
12 |
0.4 |
CGA |
Arg |
4 |
0.1 |
2 |
< 0.1 |
|
CCG |
Pro |
10 |
0.3 |
6 |
0.2 |
CGG |
Arg |
6 |
0.2 |
7 |
0.2 |
|
ACT |
Thr |
67 |
2.0 |
71 |
2.1 |
AGT |
Ser |
80 |
2.4 |
96 |
2.9 |
|
ACC |
Thr |
13 |
0.4 |
16 |
0.5 |
AGC |
Ser |
18 |
0.5 |
9 |
0.3 |
|
ACA |
Thr |
12 |
0.4 |
10 |
0.3 |
AGA |
Ser |
23 |
0.7 |
15 |
0.4 |
|
ACG |
Thr |
19 |
0.6 |
17 |
0.5 |
AGG |
Ser |
27 |
0.8 |
18 |
0.5 |
|
GCT |
Ala |
70 |
2.1 |
65 |
1.9 |
GGT |
Gly |
151 |
4.5 |
144 |
4.3 |
|
GCC |
Ala |
16 |
0.5 |
21 |
0.6 |
GGC |
Gly |
15 |
0.4 |
10 |
0.3 |
|
GCA |
Ala |
7 |
0.2 |
12 |
0.4 |
GGA |
Gly |
20 |
0.6 |
26 |
0.8 |
|
GCG |
Ala |
11 |
0.3 |
7 |
0.2 |
GGG |
Gly |
79 |
2.4 |
83 |
2.5 |
Table 4.Divergences of nucleotides and amino acids of the protein-coding genes
Table 4.
|
|
Spirometra erinaceieuropaei
|
|
|
cox1
|
cox2
|
cox3
|
cob
|
atp6
|
nad1
|
nad2
|
nad3
|
nad4
|
nad4L
|
nad5
|
nad6
|
rrnL
|
rrnS
|
|
S. decipiens
|
NCa
|
9.3 |
10.4 |
12.1 |
10.9 |
13.4 |
9.8 |
13.7 |
12.6 |
14 |
11.9 |
18.1 |
18.8 |
11.2 |
6.9 |
|
AAb
|
1.9 |
3.2 |
5.6 |
4.1 |
8.2 |
6.1 |
8.6 |
6.8 |
9.4 |
2.3 |
11.9 |
14.8 |
|
|
References
- 1. Mueller JF. A repartition of the genus Diphyllobothrium. J Parasitol 1937;23:308-310.
- 2. Faust EC, Campbell HE, Kellogg CR. Morphological and biological studies on the species of Diphyllobothrium in China. Am J Hyg 1929;9:560-583.
- 3. Le TH, Blair D, McManus DP. Mitochondrial genomes of parasitic flateworms. Trends Parasitol 2002;18:206-213.
- 4. Jeon HK, Eom KS. Molecular approaches to Taenia asiatica. Korean J Parasitol 2013;51:1-8.
- 5. Park JK, Kim KH, Kang SH, Jeon HK, Kim JH, Littlewood DTJ, Eom KS. Characterization of the mitochondrial genome of Diphyllobothrium latum (Cestoda: Pseudophyllidea)-implications for the phylogeny of eucestodes. Parasitology 2007;134:749-759.
- 6. Kim KH, Jeon HK, Kang S, Sultana T, Kim GJ, Eom KS, Park JK. Characterization of the complete mitochondrial genome of Diphyllobothrium nihonkaiense (Diphyllobothriidae: Cestoda), and development of molecular markers for differentiating fish tapeworms. Mol Cells 2007;23:379-390.
- 7. Liu GH, Li C, Yuan J, Zhou DH, Xiong RC, Lin RQ, Zou FC, Zhu XQ. Characterization of the complete mitochondrial genome sequence of Spirometra erinaceieuropaei (Cestoda: Diphyllobothriidae) from China. Int J Biol Sci 2012;8:640-649.
- 8. Zhu XQ, Beveridge I, Berger L, Barton D, Gasser RB. Singlestrand conformation polymorphism-based analysis reveals genetic variation within Spirometra erinacei (Cestoda: Pseudophyllidea) from Australia. Mol Cell Probes 2002;16:159-165.
- 9. Liu W, Liu GH, Li F, He DS, Wang T, Sheng XF, Zeng DL, Yang FF, Liu Y. Sequence variability in three mitochondrial DNA regions of Spirometra erinaceieuropaei spargana of human and animal health significance. J Helminthol 2011;1-5.
- 10. Kokaze A, Miyadera H, Kita K, Machinnami R, Noya O, de Noya BA, Okamoto M, Horii T, Kojima S. Phylogenetic identification of Sparganum proliferum as a pseudophyllidean cestode. Parasitol Int 1997;46:271-279.
- 11. Jeon HK, Kim KH, Eom KS. Complete sequence of the mitochondrial genome of Taenia saginata: comparison with T. solium and T. asiatica. Parasitol Int 2007;56:243-246.
- 12. Lowe T, Eddy SR. tRNAscan-SE: a program improved detection of transfer DNA genes in genomic sequence. Nuclei Acids Res 1997;25:955-964.
- 13. Matzura O, Wennborg A. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput Appl Biosci (CABIOS) 1996;12:247-249.
- 14. Swofford DL. Paup*: phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, Massachusetts, USA. Sinauer Associates; 2003.
- 15. Huelsenbeck JP, Ronquist F. MPBAYES: Bayesian inference of phylogenetic tree. Bioinformatics 2001;8:754-755.
- 16. Jeon HK, Park HS, Lee DM, Choe SJ, Kim KH, Huh S, Sohn WM, Chai JY, Eom KS. Human infections with Spirometra decipiens plerocercoids identified by morphologic and genetic analyses in Korea. Korean J Parasitol 2015;53:299-305.