Abstract
Protein arginine methyltransferase (PRMT) is an important epigenetic regulator in eukaryotic cells. During encystation, an essential process for Acanthamoeba survival, the expression of a lot of genes involved in the encystation process has to be regulated in order to be induced or inhibited. However, the regulation mechanism of these genes is yet unknown. In this study, the full-length 1,059 bp cDNA sequence of Acanthamoeba castellanii PRMT1 (AcPRMT1) was cloned for the first time. The AcPRMT1 protein comprised of 352 amino acids with a SAM-dependent methyltransferase PRMT-type domain. The expression level of AcPRMT1 was highly increased during encystation of A. castellanii. The EGFP-AcPRMT1 fusion protein was distributed over the cytoplasm, but it was mainly localized in the nucleus of Acanthamoeba. Knock down of AcPRMT1 by synthetic siRNA with a complementary sequence failed to form mature cysts. These findings suggested that AcPRMT1 plays a critical role in the regulation of encystation of A. castellanii. The target gene of AcPRMT1 regulation and the detailed mechanisms need to be investigated by further studies.
-
Key words: Acanthamoeba castellanii, characterization, encystation, protein arginine methyltransferase 1
INTRODUCTION
Acanthamoeba is the causative agent of granulomatous amoebic encephalitis and amoebic keratitis [
1]. To transform into the resistant cyst stage of
Acanthamoeba, the expression levels of a lot of genes mediating the encystation process have to be regulated during encystation. The comparison of transcriptome between cysts and trophozoites in
Acanthamoeba castellanii using cDNA microarray demonstrated that 701 genes were highly expressed more than 2-folds in cysts, and 859 genes were expressed lower in cysts than in trophozoites [
2]. However, the regulation mechanisms of the encystation mediating genes in
Acanthamoeba are still unknown. Therefore, here we focused on epigenetic-based regulation of gene expression during encystation of
Acanthamoeba.
Epigenetic regulation of gene expression is associated with DNA methylation, histone modification, and nucleosome positioning [
3]. Histone modification, such as methylation, acetylation, and phosphorylation, affects the state of chromatin and gene expression [
4]. Arginine methylation is a post-translational modification occurring in several cytoplasmic and nuclear proteins that modulates chromatin modification, RNA processing, DNA repair, organelle biogenesis, and signaling [
5,
6].
Protein arginine methyltransferases (PRMTs) transfer the methyl group from S-adenosylmethionine to arginine residues of proteins, and generate monomethyl arginine, symmetric dimethylarginine, or asymmetric dimethylarginine [
7]. In humans, 9 PRMTs (PRMT1 to PRMT9) have been identified, and they are classified in 4 types (I to IV) according to the modification that they generate [
6]. PRMT1 is the predominant member of the PRMT family [
8]. PRMT1 is a type I catalytic enzyme, and it is a histone methyltransferase that methylates arginine 3 on histone H4 which plays a role in activation of gene transcription [
9,
10]. PRMT1 is also a coactivator of some nuclear receptors as well as transcription factors [
11]. Coactivator-associated arginine methyltransferase 1 (CARM1) belongs to a family of previously identified PRMTs, and CARM1 can methylate arginine 17 and 26 of histone H3 [
12].
In parasitic protozoa, PRMT1 has been identified in
Plasmodium berghei (GenBank accession no. LK023125),
Plasmodium falciparum (no. SBT78822), and
Entamoeba histolytica (no. KB823531). However, the functions of PRMT1 in parasitic protozoa remain unknown. PRMT1 of
P. falciparum methylated only arginine 3 of histone H4 [
13], and PRMT1 of
E. histolytica dimethylated arginine 8 of histone H4 [
14].
In this study, we report the identification and characterization of the PRMT1 homologue in A. castellanii (AcPRMT1) (no. KT345168). For this purpose, we cloned and expressed the protein PRMT1 from our existing cDNA library of A. castellanii and investigated its role in encystation of Acanthamoeba.
MATERIALS AND METHODS
Cultivation of Acanthamoeba trophozoites and cysts
A. castellanii Castellanii (ATCC 30011) was obtained from the American Type Culture Collection. Trophozoites were axenically cultured in PYG media, consisting of proteose peptone 2% (w/v), yeast extract 0.1% (w/v), and glucose 100 mM (w/v), at 25°C in a Sanyo incubator (San Diego, California, USA). Cysts were induced in encystation media (0.1 M KCl, 0.008 M MgSO
4, 0.0004 M CaCl
2, and 0.02 M 2-amino-2-methyl-1,3-propanediol, pH 9.0) for 3 days [
15]. Mature cysts were counted under a light microscope after treating them with 0.5% SDS, and encystation ratios were calculated [
16].
Gene expression analysis
Total RNAs from
A. castellanii trophozoites and cysts were purified using RNeasy Mini Kit (Qiagen, Hilden, Germany), and cDNA synthesis was conducted using First Strand cDNA Synthesis Kit (Sigma-Aldrich, Fluka, Zwijndrecht, Netherlands) according to the manufacturer’s recommendations. Real-time PCR was carried out using 20 ng of cDNA as template and specific primers corresponding to the AcPRMT1 and AcPRMT5 genes (
Table 1). It was performed using the ABI PRISM
® 7000 sequence detection system (Applied Biosystems, Foster City, California, USA), and all reaction mixtures used SYBR Premix Ex Taq (Takara, Otsu, Shiga, Japan). The 18S ribosomal DNA was used as the reference gene [
17]. Real-time PCR was performed to determine relative gene expression using the 2
ΔΔCT method [
18], and experiments were performed in triplicate.
To investigate the intracellular localization of PRMT1, the gene was cloned into the pGAPDH vector using enhanced green fluorescent protein (EGFP) as a marker [
19]. The pGAPDHgPRMT1 plasmid was transfected into live cells of
A. castellanii. Stable transfection was performed using the SuperFect transfection reagent (Qiagen) following the manufacturer’s recommendations. Transfected cells were transferred to 50 μg neomycin-containing G418 media and grown over several passages. The amoeba expressing EGFP was allowed to adhere to a cell culture dish, and the LSM 5 EXCITER Scalable confocal system (ZEISS, Hamburg, Germany) was used to observe the amoeba. EGFP- or DAPI (4′,6-diamidino-2-phenylindole) mediated fluorescence was performed using bandpass filters covering excitation and emission wavelengths of 500 to 530 nm and 360 to 460 nm, respectively.
Small interfering RNA (siRNA) targeting PRMT1 of
A. castellanii was synthesized by Sigma-Proligo (Boulder, Colorado, USA), based on its cDNA sequence (
Table 1). The siRNA designed against AcPRMT1 (final concentration of 100 nM) was transfected into live
A. castellanii trophozoites at a cell density of 4×10
5 per well using the SuperFect transfection reagent (Qiagen) following the manufacturer’s protocol.
Data are expressed as the mean±SEM. Statistical significance was analyzed using an unpaired Student’s t-test. A P-value of <0.01 was interpreted as statistically significant.
RESULTS
Identification of AcPRMT1
To identify the epigenetic regulator in
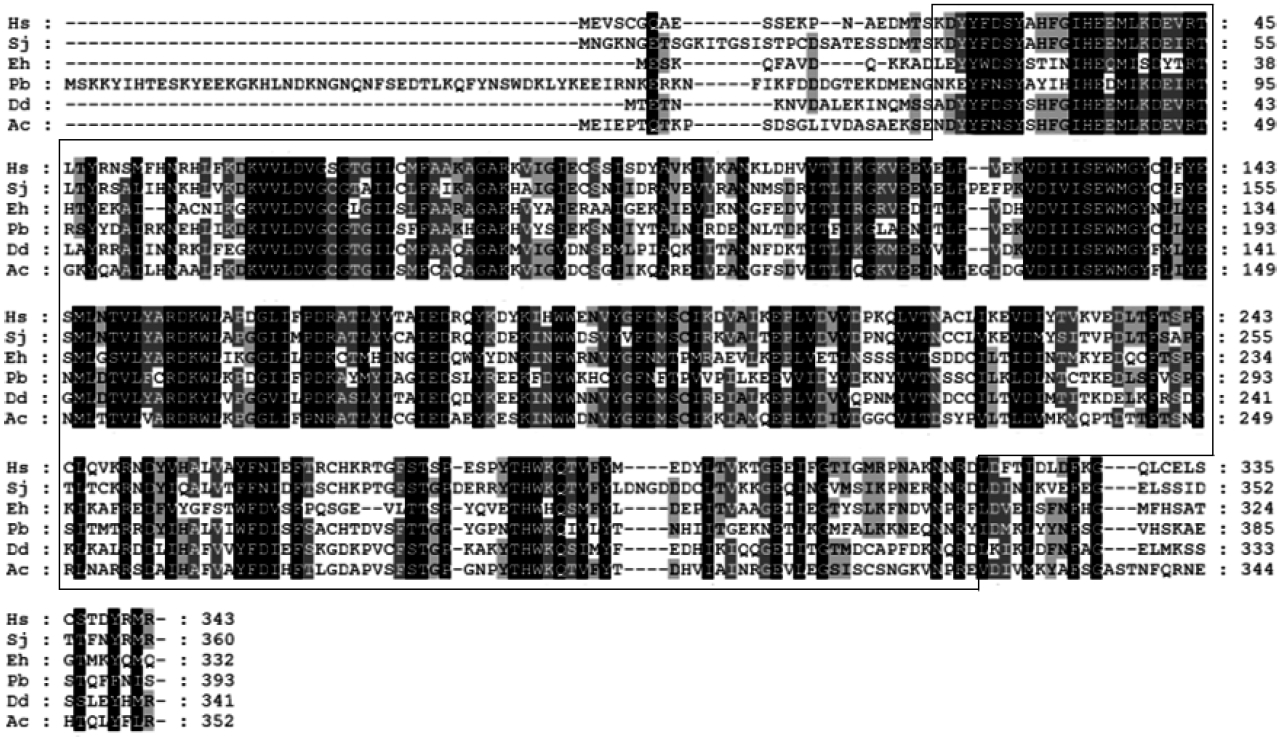
A. castellanii, we used the basic local alignment search tool (BLAST) and KOG (euKaryotic Orthologous Groups) analysis from our existing cDNA library, and identified a full-length open reading frame of AcPRMT1 (GenBank accession no. KT345168). AcPRMT1 has 352 amino acids with a predicted molecular mass of 38.7 kDa. Based on homology searches, the deduced amino acid sequences of the AcPRMT1 cDNA showed 42.6% similarity with AcPRMT5 (GenBank accession no. KT345169) (data not shown). Amino acid sequence alignment of AcPRMT1 with that of
Schistosoma japonicum,
E. histolytica,
Dictyostelium discoideum,
P. berghei, and
Homo sapiens showed sequence similarity (
Fig. 1). Putative AcPRMT1 encodes a SAM-dependent methyltransferase PRMT-type domain (boxed area).
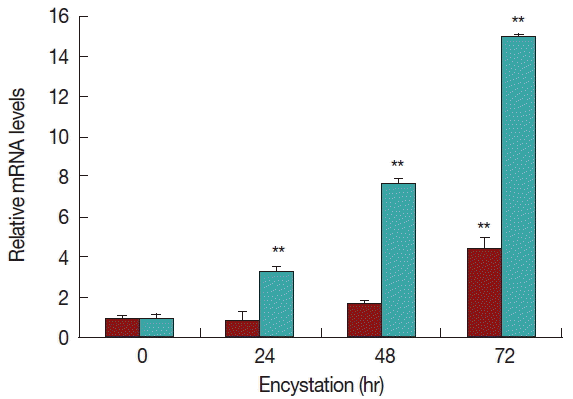
Real-time PCR analysis was used to detect the transcript levels of AcPRMT1 during encystation of
A. castellanii. The expression levels of AcPRMT1 were highly increased after induction of encystation from 24–72 hr (
Fig. 2). The previously identified PRMT5 of
A. castellanii (AcPRMT5) also showed increased expression patterns during encystation [
19]. However, AcPRMT1 was expressed higher than AcPRMT5 during encystation of
A. castellanii (
Fig. 2).
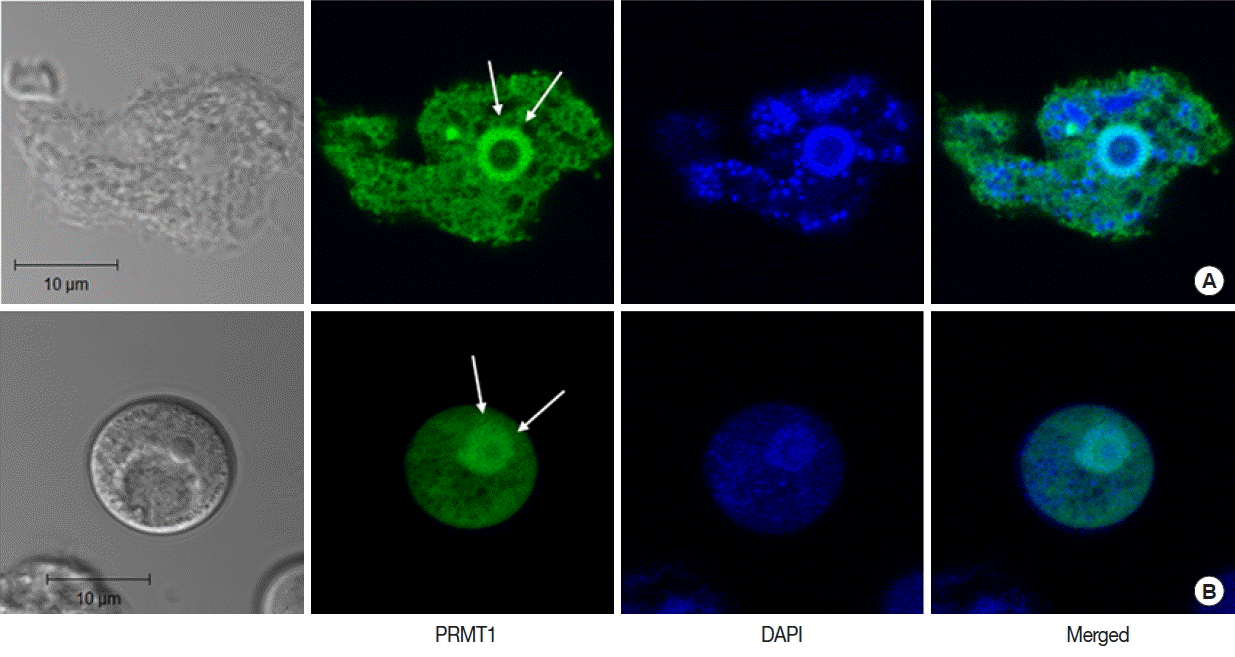
In
A. castellanii transfected with EGFP-tagged AcPRMT1, strong GFP fluorescence was detected in the cytoplasm and the nucleus (
Fig. 3). Previously it was found that the EGFP-AcPRMT5 fusion protein was mainly localized in the nucleus of
Acanthamoeba [
19]. However, AcPRMT1 was localized in both the cytoplasm and the nucleus of
Acanthamoeba trophozoite (
Fig. 3A) and cyst (
Fig. 3B). The localization of AcPRMT1 in the nucleus was confirmed by DAPI staining (
Fig. 3).
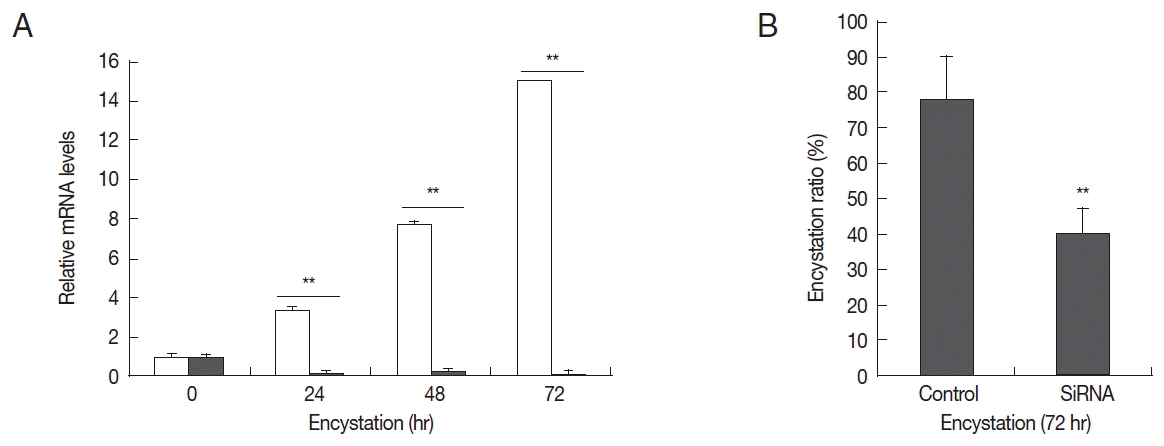
To determine the effects of AcPRMT1 on encystation of
A. castellanii, post-transcriptional gene silencing by siRNAs was performed. After transfection of fluorescent tagged siRNA against AcPRMT1 (
Table 1), the transfection efficiency of siRNA was determined to be 80% by fluorescent microscopy. Transfected trophozoites were transferred to the encystment media, and the effect of AcPRMT1 gene silencing on encystation of
Acanthamoeba was investigated. As shown in
Fig. 4A, the mRNA expression of AcPRMT1 was successfully decreased during encystation. Decreased expression of AcPRMT1 by siRNA affected the encystation ratio of
Acanthamoeba. The number of mature cysts was significantly reduced in siRNA transfected cells (40.2%) compared to control cells (78.4%) (
Fig. 4B).
DISCUSSION
Histone arginine methylation by PRMTs has been shown to play an important role in transcriptional regulation [
20,
21]. In this study, PRMT1 was identified for the first time in
A. castellanii (AcPRMT1). We suggested that PRMT1 play an important role in encystation of
Acanthamoeba based on the result that the cells transfected with siRNA against AcPRMT1 failed to form mature cysts (
Fig. 4). AcPRMT1 was localized both in the cytoplasm and the nucleus (
Fig. 3), while we have previously identified that AcPRMT5 was localized mainly in the nucleus [
19]. However, the substrates of PRMT1 and PRMT5 of
A. castellanii have yet to be identified. PRMT1 of
P. falciparum was localized in both the cytoplasm and the nucleus, and this suggested the presence of potential substrates in both cellular compartments [
13].
PfPRMT1 methylated histone H4 predominant at arginine 3 and formed monomethylarginine (MMA) and asymmetric dimethylarginine (ADMA) [
13]. The authors also previously confirmed the presence of histone H3 arginine 17 methylation in
P. falciparum, a potential substrate for the CARM-like PRMT. PRMT1 of
S. mansoni was able to specifically methylate histone H4, but not histone H3 [
22]. Unfortunately, the sequence of histone H4 of
A. castellanii Castellani has not yet been reported. Also, histone H4 arginine 3 position in
H. sapiens was different from that in
A. castellanii Neff (GenBank accession no. ELR24196). However, histone H3 arginine residues (Arg
2, Arg
8, and Arg
17) of
A. castellanii Castellani and
A. castellanii Neff (no. ELR19918) are the same with those of humans (no. AAN39284) (data not shown). It is highly supposed that AcPRMT1 is able to methylate histone H3 arginine 17 of
Acanthamoeba (MARTKQTARKSTGGKAP
RKMAS) similar to CARM-like PRMT.
PRMT1 mediated dimethylation of histone H4 arginine 3 is associated with gene activation [
23], while the methylation of histone H4 arginine 3 by PRMT5 is associated with gene repression [
24]. The authors hypothesized that AcPRMT1 and AcPRMT5 regulate the transcriptional levels of encystation associated genes by histone modification. Furthermore, we hypothesized that AcPRMT1 is associated with gene expression, while AcPRMT5 is associated with gene repression. AcPRMT1 was expressed higher than AcPRMT5 during encystation (
Fig. 2), which suggested that there are various active genes associated with encystation.
On the basis of these results, further research is required to obtain target substrates and specific inhibitors of AcPRMT1. We predict that PRMT1 plays a key role in the expression of certain resistance genes in encysting Acanthamoeba. Further research will lead to the development of anti-amoebic drugs for human Acanthamoeba infections.
Notes
-
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant no. 2014R1A1A 2058405), and Kyung Hee University in 2016 (no. KHU 20160 537).
Fig. 1Alignment of the PRMT1 amino acid sequences of A. castellanii (GenBank accession no. KT345168) with that of the sequences of H. sapiens (no. BC109282), S. japonicum (no. FN318749), E. histolytica (no. KB823531), P. berghei (no. LK023125), and D. discoideum (no. Q54EF2). Identical and similar amino acids are shaded in black or grey, respectively. Boxed area shows a SAM-dependent methyltransferase PRMT-type domain.
Fig. 2Expression of AcPRMT1 and AcPRMT5 during encystation. The mRNA expression levels of AcPRMT1 and AcPRMT5 were highly increased during encystation of A. castellanii in a time-dependent manner. Expression levels of PRMT1 (green bars) were higher than those of PRMT5 (red bars). **The means are significantly different at P<0.01 by Student’s t-test.

Fig. 3Confocal microscopic images of A. castellanii with stable transfection of pGAPDHgPRMT1 for intracellular localization of AcPRMT1. The EGFP-AcPRMT1 fusion protein was distributed over the cytoplasm but it was mainly localized in the nucleus (arrows). Localization of AcPRMT1 in the nucleus was confirmed by DAPI staining. (A) Trophozoite, (B) Cyst.
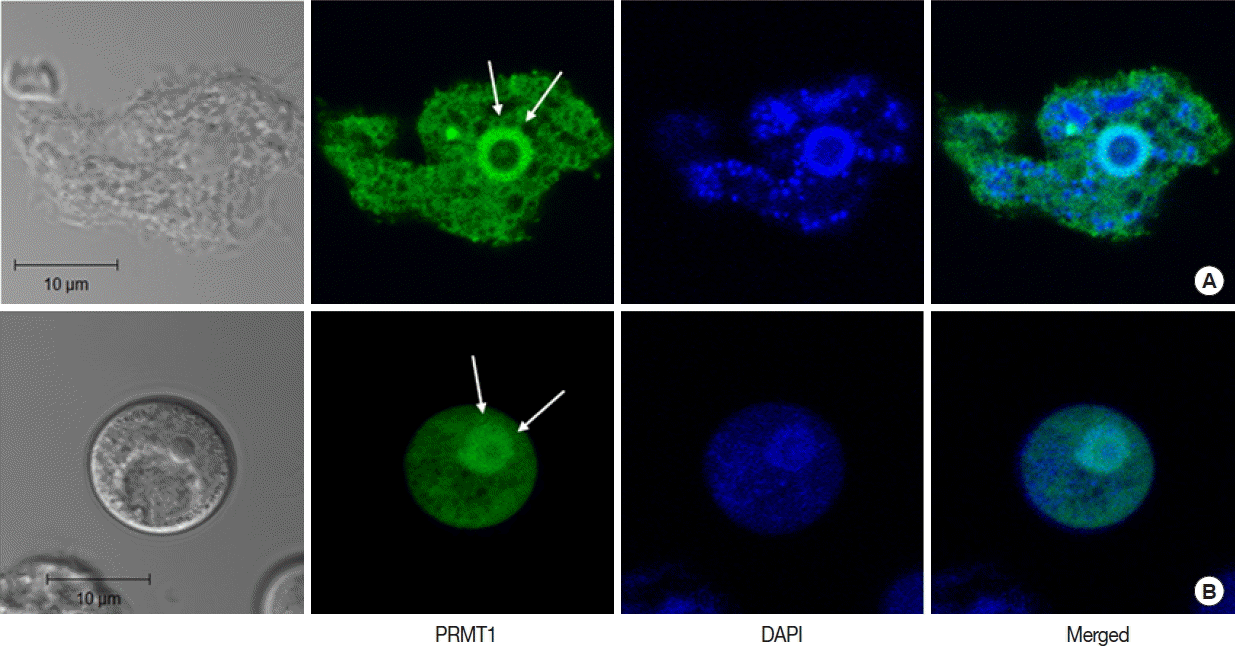
Fig. 4Expression levels of AcPRMT1 and inhibition of encystation by AcPRMT1-siRNA. (A) Expression levels of AcPRMT1. The expression of the AcPRMT1 gene during encystation was almost completely inhibited by AcPRMT1-siRNA treatment (close square) compared to the control (open square). (B) Inhibition of encystation. The AcPRMT1-siRNA transfected cells showed reduced encystation ratios than the control cells. Values indicate the mean±SD of 3 experiments. **Means are significantly different (Student’s t-test, P<0.01).
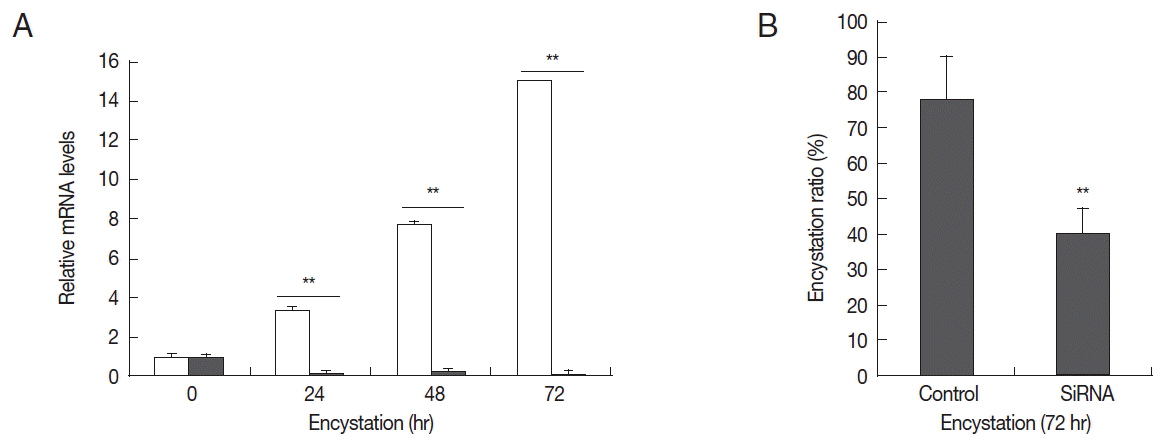
Table 1Primer sequences used in the study
Table 1
|
Primer |
Sequence (5′-3′) |
|
PRMT1-real time-F |
TTCCCCAACAGAGCTACCCTCT |
|
PRMT1-real time-R |
GCAGCTCATGTCAAAGCCGTA |
|
PRMT5-real time–F |
TTGACTACTCGGCTCTTCTGCC |
|
PRMT5-real time-R |
TCGATGTCTTTCACCAGCAGG |
|
PRMT1-pGAPDHgx-F |
ACATCTAGAATGGAAATCGAACCGACTCA |
|
PRMT1-pGAPDHgx-R |
ATATCTAGATTAGCGGAGGAAGTAGAGCT |
|
PRMT1-siRNA-F |
CCUACUUCGACAUUCACUUdTdT |
|
PRMT1-siRNA-R |
AAGUGAAUGUCGAAGUAGGdTdT |
References
- 1. Marciano-Cabral F, Cabral G. Acanthamoeba spp. as agents of disease in humans. Clin Microbiol Rev 2003;16:273-307.
- 2. Moon EK, Xuan YH, Chung DI, Hong Y, Kong HH. Microarray analysis of differentially expressed genes between cysts and trophozoites of Acanthamoeba castellanii. Korean J Parasitol 2011;49:341-347.
- 3. Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol 2010;28:1057-1068.
- 4. Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bähler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 2007;449:928-932.
- 5. Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell 2009;33:1-13.
- 6. Pahlich S, Zakaryan RP, Gehring H. Protein arginine methylation: cellular functions and methods of analysis. Biochim Biophys Acta 2006;1764:1890-1903.
- 7. Niewmierzycka A, Clarke S. S-adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J Biol Chem 1999;274:814-824.
- 8. Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol 2000;20:4859-4869.
- 9. Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther 2007;113:50-87.
- 10. Katsanis N, Yaspo ML, Fisher EM. Identification and mapping of a novel human gene, HRMT1L1, homologous to the rat protein arginine N-methyltransferase 1 (PRMT1) gene. Mamm Genome 1997;8:526-529.
- 11. Nicholson TB, Chen T, Richard S. The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation. Pharmacol Res 2009;60:466-474.
- 12. Kraus WL, Wong J. Nuclear receptor-dependent transcription with chromatin. Is it all about enzymes? Eur J Biochem 2002;269:2275-2283.
- 13. Fan Q, Miao J, Cui L, Cui L. Characterization of PRMT1 from Plasmodium falciparum. Biochem J 2009;421:107-118.
- 14. Borbolla-Vázquez J, Orozco E, Betanzos A, Rodríguez MA. Entamoeba histolytica: protein arginine transferase 1a methylates arginine residues and potentially modify the H4 histone. Parasit Vectors 2015;8:219.
- 15. Bowers B, Korn ED. The fine structure of Acanthamoeba castellanii (Neff strain). II. Encystment. J Cell Biol 1969;41:786-805.
- 16. Aqeel Y, Siddiqui R, Khan NA. Silencing of xylose isomerase and cellulose synthase by siRNA inhibits encystation in Acanthamoeba castellanii. Parasitol Res 2013;112:1221-1127.
- 17. Moon EK, Hong Y, Chung DI, Kong HH. Cysteine protease involving in autophagosomal degradation of mitochondria during encystation of Acanthamoeba. Mol Biochem Parasitol 2012;185:121-126.
- 18. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001;25:402-408.
- 19. Moon EK, Hong Y, Chung DI, Goo YK, Kong HH. Identification of protein arginine methyltransferase 5 as a regulator for encystation of Acanthamoeba. Korean J Parasitol 2016;54:133-1338.
- 20. Litt M, Qiu Y, Huang S. Histone arginine methylations: their roles in chromatin dynamics and transcriptional regulation. Biosci Rep 2009;29:131-141.
- 21. Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett 2011;585:2024-2031.
- 22. Mansure JJ, Furtado DR, de Oliveira FM, Rumjanek FD, Franco GR, Fantappié MR. Cloning of a protein arginine methyltransferase PRMT1 homologue from Schistosoma mansoni: evidence for roles in nuclear receptor signaling and RNA metabolism. Biochem Biophys Res Commun 2005;335:1163-1172.
- 23. Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 2001;293:853-857.
- 24. Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 2004;24:9630-9645.