Abstract
The present study was performed with morphological and molecular analysis (cox1 and nad1 mitochondrial genes) to identify the proglottids of spirometrid tapeworm found in the stool of an African lion, Panthera leo, in the Serengeti plain of Tanzania. A strand of tapeworm strobila, about 75 cm in length, was obtained in the stool of a male African lion in the Serengeti National Park (34° 50′ E, 02° 30′ S), Tanzania, in February 2012. The morphological features of the adult worm examined exhibited 3 uterine coils with a bow tie appearance and adopted a diagonal direction in the second turn. The posterior uterine coils are larger than terminal uterine ball and the feature of uteri are swirling rather than spirally coiling. The sequence difference between the Spirometra species (Tanzania origin) and S. erinaceieuropaei (GenBank no. KJ599680) was 9.4% while those of S. decipiens (GenBank no. KJ599679) differed by 2.1% in the cox1 and nad1 genes. Phylogenetic tree topologies generated using the 2 analytic methods were identical and presented high level of confidence values for the 3 major branches of the 3 Spirometra species in the cox1 gene. The morphological and molecular findings obtained in this study were nearly coincided with those of S. ranarum. Therefore, we can know for the first time that the African lion, Panthera leo, is to the definitive host of this tapeworm.
-
Key words: Spirometra ranarum, cox1, nad1, lion, Tanzania
Tapeworms in the genus
Spirometra (Cestoda: Diphyllobothriidae) are mainly reside in the small intestines of cats and dogs. They are morphologically divided into 6 species, i.e.,
S. erinaceieuropaei (Rudolphi, 1819),
S. decipiens (Diesing, 1850),
S. ranarum (Gastaldi, 1854),
S. mansoni (Cobbold, 1882)
S. houghtoni (Syn.
S. mansoni, Faust et al., 1929) and
S. okumurai (Faust et al., 1929) by Faust et al. [
1]. In the American continent, a total of 5 species, i.e.,
Spirometra species,
S. decipiens,
S. mansoni,
S. gracilis (Baer, 1927),
S. longicollis (Parodi and Widakowich, 1917) and
S. mansonoides (Mueller, 1935), have been reported in the literatures [
2,
3]. However, Kamo [
4] acknowledged only 4 species, i.e.,
Spirometra species,
S. erinaceieuropaei,
S. pretoriensis (Baer, 1924),
S. theileri (1924) and
S. mansonoides as valid species.
In African continent, domestic dogs and cats are to be known as the main definitive hosts of spirometrid tapeworms and some species of wild carnivores are also reported as the final hosts of
Spirometra tapeworms [
5]. A
Spirometra species from African mammals was first described by Baer (1925) [
6]. Egg positive rates of
Spirometra sp. were reported in the stool examinations of hyenas in Kenya (74%) and Zambia (22%), and in lions in Tanzania (63%) and Zambia (87%) [
7–
11]. Plerocercoid larvae of
Spirometra spp. (spargana) were detected in wild herbivores, i.e., antelope, buffalo, zebra, wildebeest and wart hogs, in Tanzania and Kenya [
9–
11], and found in Baboons, vervets and other monkeys in East Africa [
12,
13]. On the other hand, some infection cases with spirometrid tapeworms in lions were previously reported in Africa [
9–
12]. Two valid species,
S. theileri and
S. pretoriensis, have been confirmed to infect lions as well as hyenas [
6]. However, it is very obscure that what other species of
Spirometra except for 2 species exist in the ecosystems of these tapeworms in Africa. Accordingly, it is needed to identify the larvae and adults of
Spirometra spp. from various sources of animals, the second intermediate or paratenic and definitive hosts including lions.
In this study, a Spirometra species of Tanzania origin from lion was identified by molecular analysis of 2 complete mitochondrial genes, cytochrome c oxidase I (cox1) and NADH dehydrogenase subunit 1 (nad1), as well as by morphological observations of an adult tapeworm obtained from an African male lion.
One strobila of a
Spirometra species was obtained in the stool of male African lion (
Panthera leo) in Seronera (at a research station in the center of the Serengeti National Park, 34° 50′ E, 02° 30′ S), Tanzania, East of Africa in February 2012. The collected worm was fixed in 70% ethanol and some gravid proglottids were compressed between 2 glass slides prior to fixation in 10% neutral buffered formalin. Microscopic observations were then performed after carmine staining. The morphologic data were compared to those of Faust et al. [
1] with emphasis on the vaginal opening, uterus, uterine pore, cirrus, genital pore, testes, and vitellaria of gravid proglottids.
Some of the proglottids in 70% ethanol were used for molecular studies. Protocols employed for total genomic DNA extraction and PCR reactions were those previously described by Jeon et al. [
14]. The entire
cox1 and
nad1 genes were amplified by PCR. The complete sequence of the mitochondrial
cox1 gene was amplified using primers Spi-CO1F: 5′-GAC TAA GTG TTT TCA AAA CAC TAA GTG -3′ and Spi-CO1R: 5′CAC CCT ACC CCT GAT TTA CAA AAT-3′, and for cycle sequencing of the PCR product one internal primer, Spi-Ci1R2: 5′-CCA ATA ATC ATG GTC ACA GAA C-3′, was used in addition to the PCR primers to obtain the complete sequence. The complete sequence of the mitochondrial
nad1 gene was amplified using primers Spi-ND1F: 5′-GGA GAA TAT TGG TTT GTC TAA CCA- 3′ and Spi-ND1R: 5′-CCT TCT TAA CGT TAA CAG CAT TAC GAT-3′, which were designed from the complete sequences of
S. erinaceieuropaei (KJ599680) and
S. decipiens (KJ599679) mitochondrial genomes and used to amplifying the complete
cox1 and
nad1 genes, respectively.
The DNA sequences were assembled and aligned using the Geneious 9.0 program (Biometer, Auckland, New Zealand). These sequences were identified by comparison with sequences of
S. erinaceieuropaei and
S. decipiens in the GenBank database. Phylogenetic analysis was evaluated using Bayesian inference (BI) and maximum-likelihood (ML) using the complete mitochondrial
cox1 (1,566 bp) and
nad1 (891 bp) sequences of
S. erinaceieuropaei (KJ599680),
S. decipiens (KJ599679), and
S. ranarum (MH298843, MH298844). The following complete mitochondrial
cox1 sequences were used:
S. erinaceieuropaei (KJ 599680),
S. decipiens (KJ599679, LC328893-328901, LC213623, AB369249, AB374543),
S. ranarum (AB369250-369251, MH298843),
Diphyllobothrium latum (DQ985706) and
D. nihonkaiense (EF420138). ML analyses of
cox1 and
nad1 used RAxML v. 7.3.1 [
15] after GTR+G+I substitution model sampling was chosen according to the Modeltest using the program Partition Finder [
16]. BI analyses were used in MrBayes 3.2 and running 4 simultaneous Monte Carlo Markov chains (MCMC) for 10 million generations and discarding the first 25% generation as burn-in every 1,000 generations [
17].
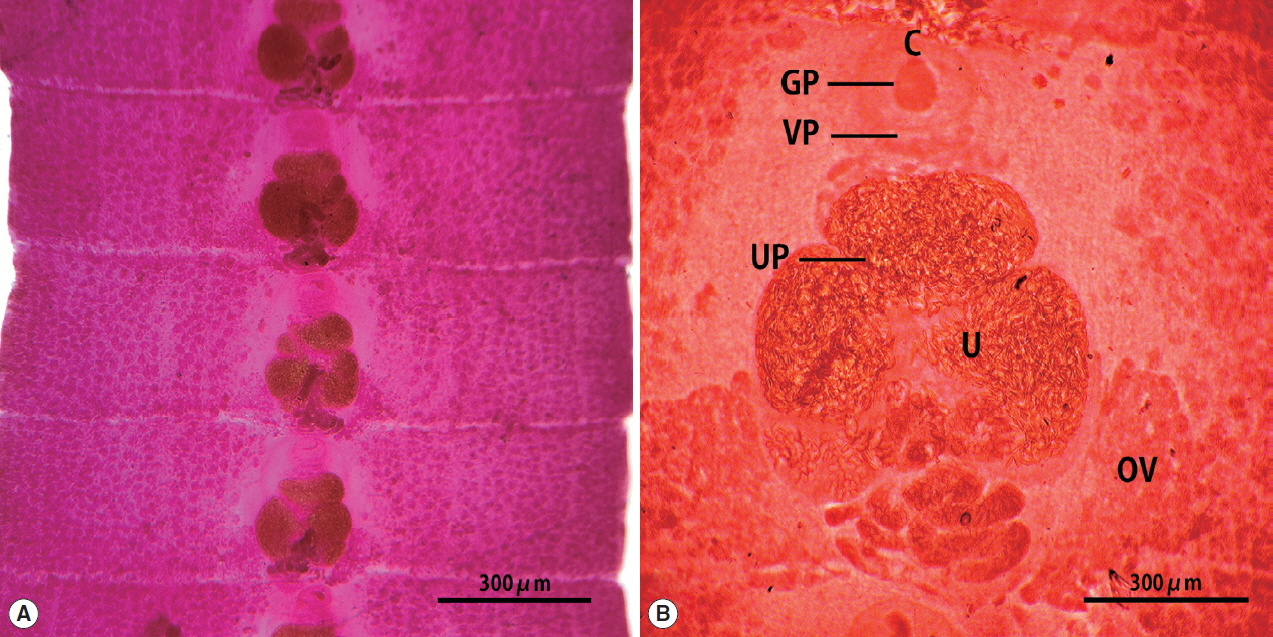
The strobila collected from the stool of a lion was 75 cm. The mature and gravid proglottids were 2.5×12.0 mm in average size. The genital and vaginal pore were located ventrally on the midline in the anterior 1/3 of the segment. The uterine pore was situated on the midline behind the anterior margin of the terminal uterine ball and the terminal uterine ball measured 370 μm (n=10) in diameter. The uterus consisted of 3 loops with a bow tie appearance and adopted a diagonal direction in the second turn. The testes measured 74 μm (n=10) in diameter. The ovary was highly dendritic and connected to the uterus (
Fig. 1).
The similarity of
cox1 sequences (1,566 bp) of the
Spirometra species (Tanzania origin) was 99.7% to the reference sequences of
Spirometra species (Myanmar origin, GenBank no. MH298843) which was identified as
S. ranarum. The similarity to other
Spirometra species was 97.9% (
S. decipiens) and 90.6% (
S. erinaceieuropaei). The similarity of the
nad1 sequence (891 bp) of
Spirometra species (Tanzania origin) to reference sequences was 97.9% (
S. decipiens) and 90.6% (
S. erinaceieuropaei). The
cox1 sequence (1,566 bp) revealed 33 polymorphic sites, with 7 non-synonymous and 26 synonymous substitutions, while the
nad1 sequence (891 bp) showed 19 polymorphic sites, with 3 non-synonymous and 16 synonymous substitutions between the samples of present study and
S. decipiens (GenBank no. KJ599679) (
Table 1). The sequence difference between the
Spirometra species (Tanzania origin) and
S. erinaceieuropaei (GenBank no. KJ599680) was 9.4% while those of
S. decipiens (GenBank no. KJ599679) differed by 2.1% in the
cox1 gene. Phylogenetic tree topologies generated using the 2 analytic methods were identical and presented high level of confidence values for the 3 major branches of the 3
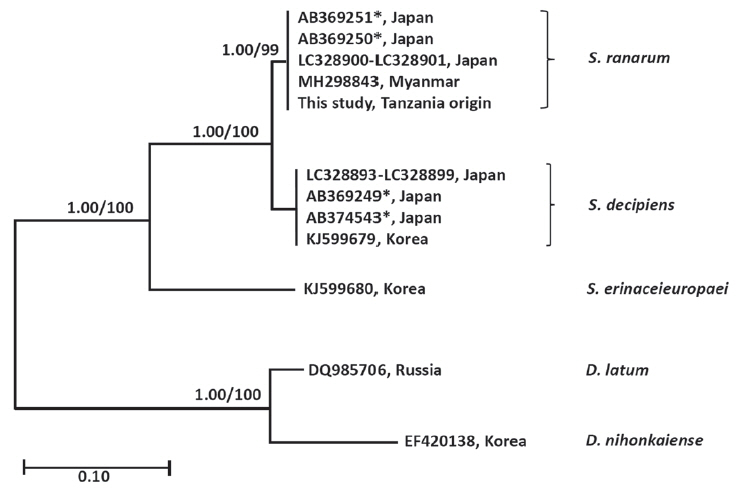
Spirometra species in the
cox1 gene. Species of
Spirometra were divided into 2 clades containing
S. erinaceieuropaei and
S. decipiens, and the
S. decipiens sub-clades including
S. decipiens and its sister species,
S. ranarum (
Fig. 2).
The cestode parasites of the African lion (
Panthera leo) were reported to comprise
Taenia regis, T. gonyamai, T. simbae, Echinococcus sp., and
Spirometra species from free-ranging wild lions of Eastern Africa [
10]. The
Spirometra species was the most prevalent parasite with the highest number of eggs in feces followed by the taeniid species [
11]. The
Spirometra species in lion is
S. theileri, whereas the species found in hyenas is commonly known as
S. pretoriensis. However, both
Spirometra species are found in lions because lions and hyenas prey on the same kind of wild herbivores such as antelope, buffalo, zebra and warthog.
In this study, some proglottids of spirometrid tapeworms obtained from the feces of lion was identified according to the methods of Faust et al. [
1] and Graber [
18]. The morphological features of our specimen were distinct from that of
S. theileri and
S. pretoriensis. The major differentiating features were the ratio of width and length of gravid proglottids and uterine morphology, with a few other features such as the uterus of
S. theileri consisting of 4–7 complete turns, while that of
S. proetoriensis consists of 2–3 coils [
18]. The morphological characteristics of
S. ranarum includes the width of gravid proglottids being larger than that of other
Spirometra spp. including
S. erinaceieuropaei,
S. decipiens,
S. theileri and
S. pretoriensis. The size of the uterine ball is also larger than that of other
Spirometra spp. Another major differentiating feature is that the uterus adopts a diagonal swirling formation rather than a spiral coil. These morphological characteristics of the Tanzania origin
Spirometra specimen in this study are consistent with those of
S. ranarum. The morphological characteristics of
S. ranarum have been detailed by Faust et al. [
1] as comprising 3–4½ uterine coils, of which the posterior 2 coils are broader and more dilated than the uterine terminal ball, highly dendritic ovaries, the testes and vitellaria are closely located to the lateral margins of the lower 2 coils of the outer uteri, the vaginal and male genital pores are located in the median plane a small distance anterior to the terminal uterine coil, and the eggs are similar in shape to those of
S. decipiens [
1].
The literature to date indicates that molecular data of mitochondrial genes provides genetic variation for taxonomical distinction and species identification for morphologically similar parasites. Recently, molecular studies for
Spirometra species were reported from many endemic areas in Asian, South American and African countries. The sequence variation of the mitochondrial
cox1 gene of
Spirometra species was 0.0–3.5% in China, Myanmar, Thailand and Lao PDR [
19]. The nucleotide sequence variation in
Spirometra spp.
cox1 twas reported as being 2.6% from Japan, India and Indonesia [
20]. This study has shown that sequence differences in the
cox1 and
nad1 genes between
Spirometra species (Tanzania origin) and
S. decipiens was 2.1%, while those of
S. erinaceieuropaei were 9.4% (
cox1) and 11.9% (
nad1), whereas those of Myanmar and Japan origin were 0.2% (
cox1). These results indicated that the molecular data of the
Spirometra specimen (Tanzania origin) are consistent with those of
S. ranarum (Myanmar origin, GenBank no. MH298843-298844).
To date the mitochondrial molecular data of Spirometra species has yet to be explained in terms of geographical distribution and human sparganosis. The findings of the present study demonstrated that Asian isolates of Spirometra species were found as S. ranarum in Myanmar, Korea and Japan by analysis of mitochondrial cox1 sequences. The sequence homologies in the cox1 gene was 99.8% between Spirometra species (Tanzania origin) and Asian isolates including China, Japan, Korea, Lao PDR, and Thailand. Thus, S. ranarum were found in China, Japan, Korea, Lao PDR and Thailand by mitochondrial cox1 sequences from GenBank database (Unpublished observation). Finally, the morphological features and molecular data of the Tanzania-origin Spirometra specimen examined coincided well with those of S. ranarum, which demonstrate the distribution of S. ranarum in Tanzania, East Africa. Therefore, we can know for the first time that the African lion, Panthera leo, is to the definitive host of this tapeworm. Further studies are needed to clarify taxonomy of the genus Spirometra including S. theileri and S. pretoriensis found in African continent.
Notes
-
CONFLICT OF INTEREST
We have no conflict of interest related to this work.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (No. 2017R1D1A3B03035976). The materials used were provided by the Parasite Resource Bank of Korea (PRB000720).
Fig. 1Gravid proglottids of Spirometra ranarum (Tanzania origin) collected from an African male lion. Whole mounted specimens of proglottids showing the cirrus (C), uterus (U), genital pore (GP), vaginal pore (VP), uterine pore (UP), and ovary (OV) (acetocarmine stain). (A)×12, (B)×40.
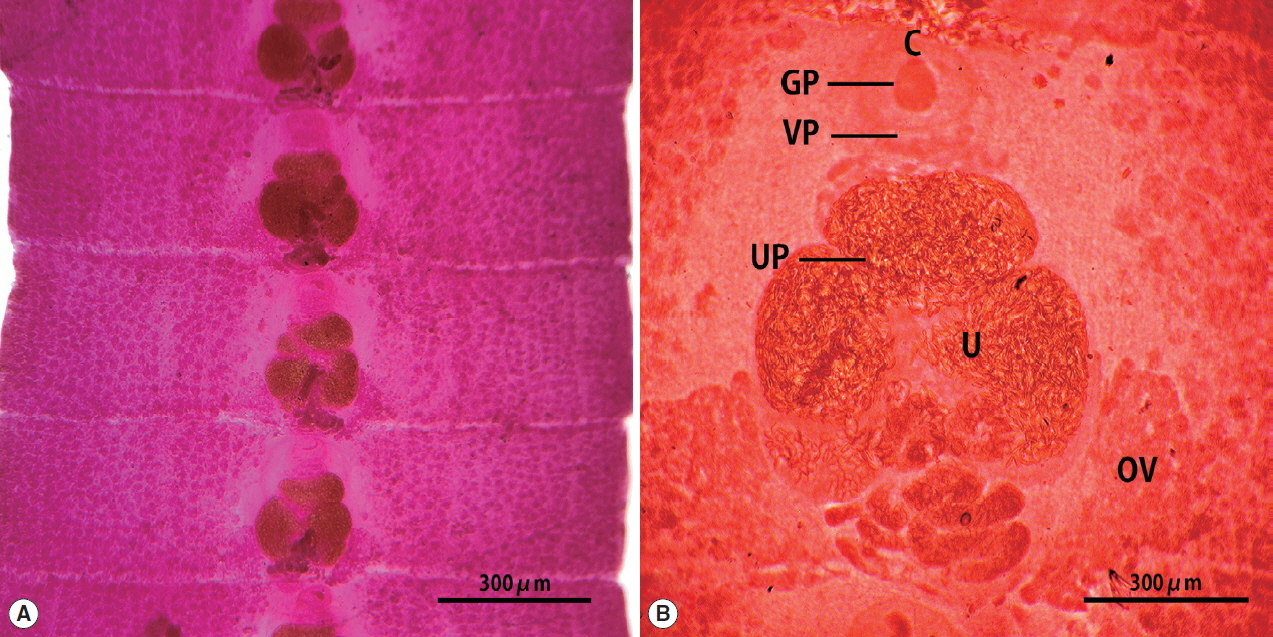
Fig. 2Phylogenetic tree of Spirometra species based on complete mitochondria cox1 sequences. Numbers above the branches represent bootstrap value for Bayesian inference (BI) and maximum likelihood (ML). *GenBank No. AB369249-AB369251, AB374543 are currently registered as S. erinaceieuropaei by complete mitochondrial cox1 sequences. GenBank No. LC328893-LC328901 are registered as S. decipiens by complete mitochondrial cox1 sequences.
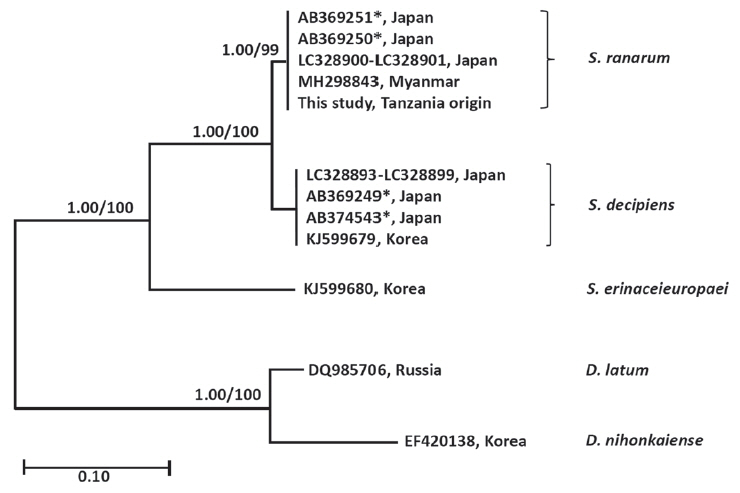
Table 1Percentage pairwise sequence homologies of the complete mitochondrial cox1 and nad1 genes of Spirometra ranarum isolated from an African lion in Tanzania and various Spirometra species
Table 1
|
Species GenBank No. |
S. erinaceieuropaei (KJ599680) |
S. decipiens (KJ599679) |
S. ranarum (MH298843, MH298844) |
|
|
|
|
cox1
|
nad1
|
cox1
|
nad1
|
cox1
|
nad1
|
|
S. ranarum*
|
90.6 |
88.1 |
97.9 |
97.9 |
99.8 |
99.8 |
References
- 1. Faust EC, Campbell HE, Kellogg CR. Morphological and biological studies on the species of Diphyllobothrium in China. Am J Epidemiol 1929;9:560-583.
- 2. McIntosh A. New Host Records for Diphyllobothrium mansonoides Mueller, 1935. J Parasitol 1937;23:313-315.
- 3. Schmidt GD. CRC Handbook of Tapeworm Identification. Boca Raton, USA. CRC Press; 1986.
- 4. Kamo H. Guide to Identification of Diphyllobothriid Cestodes. Tokyo, Japan. Gendai Kikaku; 1999, pp 1-146 (in Japanese).
- 5. Nelson GS, Pester FR, Rickman R. The significance of wild animals in the transmission of cestodes of medical importance in Kenya. Trans R Soc Trop Med Hyg 1965;59:507-524.
- 6. Baer JG. Contributions to the helminth fauna of South Africa. Mammalian Cestodes. Africa Dept Agric Rept Dir Vet Educ and Res 1926;11:63-136.
- 7. Engh AL, Nelson KG, Peebles R, Hernandez AD, Hubbard KK, Holekamp KE. Coprologic survey of parasites of spotted hyenas (Crocuta crocuta) in the Masai Mara National Reserve, Kenya. J Wildl Dis 2003;39:224-227.
- 8. Berensten AR, Becker MS, Stockdale-Walden H, Matandiko W, McRobb R, Dunbar MR. Survey of gastrointestinal parasite infection in African lion (Panthera leo), African wild dog (Lycaon pictus) and spotted hyaena (Crocuta crocuta) in the Luangwa Valley, Zambia. Afr Zool 2012;47:363-368.
- 9. Müller-Graf CD. A coprological survey of intestinal parasites of wild lions (Panthera leo) in the Serengeti and the Ngorongoro Crater, Tanzania, East Africa. J Parasitol 1995;8:812-814.
- 10. Bjork KE, Averbeck AG, Stromberg BE. Parasites and parasite stages of free-ranging wild lions (Panthera leo) of northern Tanzania. J Zoo Wild Med 2000;31:56-61.
- 11. Müller-Graf CD, Woolhouse ME, Packer C. Epidemiology of an intestinal parasite (Spirometra spp.) in two populations of African lions (Panthera leo). Parasitology 1999;118:407-415.
- 12. Kuntz RE, Myers BJ, Katzberg AA. Sparganosis and “proliferative” spargana in vervets (Cercopithecus aethiops) and baboons (Papio sp.) from East Africa. J Parasitol 1970;56:196-197.
- 13. Nobrega-Lee M, Hubbard G, Loverde P, Carvalho-Queiroz C, Conn DB, Rohde K, Dick EJ Jr, Nathanielsz P, Martin D, Siler-Khodr T, Schlabritz-Loutsevitch N. Sparganosis in wild-caught baboons (Papio cynocephalus anubis). J Med Primatol 2007;36:47-54.
- 14. Jeon HK, Park H, Lee D, Choe S, Kim KH, Huh S, Sohn WM, Chai JY, Eom KS. Human infections with Spirometra decipiens plerocercoids identified by morphologic and genetic analyses in Korea. Korean J Parasitol 2015;53:299-305.
- 15. Stamatakis A. RAxML-VI-HPC: maximum likelihood based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006;22:2688-2690.
- 16. Lanfear R, Calcott B, Ho SY, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Bio Evol 2012;29:1695-1701.
- 17. Ronquist F, Huelsenbeck JP. MrBayes 3: Baysian phylogenetic inference under mixed models. Bioinformatics 2003;9:1572-1574.
- 18. Graber M. Diphyllobothriose et sparganose en Afrique tropicale. Rev Elev Med Vet Pays Trop 1981;34:303-311.
- 19. Jongthawin J, Intapan PM, Sanpool O, Sadaow L, Laymanivong S, Thanchomnang T, Maleewong W. Molecular evidence of Spirometra erinaceieuropaei infection in snakes Ptyas korros from Lao PDR and Thailand and frogs Hoplobatrachus rugulosus from Myanmar. Southeast Asian J Trop Med Public Health 2014;45:1271-1278.
- 20. Okamoto M, Iseto S, Shibahara T, Sato MO, Wandra T, Craig PS, Ito A. Intraspecific variation of Spirometra erinaceieuropaei and phylogenetic relationship between Spirometra and Diphyllobothrium inferred from mitochondrial CO1 gene sequence. Parasitol Int 2007;56:235-238.