Abstract
Blastocystis, an enteric protist, has been reported to be an important cause of protozoal gastrointestinal manifestations in humans and animals worldwide. Animals harboring certain Blastocystis subtypes (STs) may serve as a potential source of human infection. However, information about the prevalence and genetic diversity of Blastocystis in alpacas is limited. In the present study, a total of 366 fecal samples from alpacas in Shanxi Province, northern China, were examined for Blastocystis by PCR amplification of the small subunit rRNA gene, followed by sequencing and phylogenetic analysis. The prevalence of Blastocystis in alpacas was 23.8%, and gender difference in the prevalence of Blastocystis was observed. The most predominant Blastocystis ST was ST10, followed by ST14 and ST5. The detection of ST5, a potentially zoonotic genotype, indicates that alpacas harboring ST5 could be a potential source of human infection with Blastocystis. These data provide new insight into the prevalence and genetic diversity of Blastocystis in alpacas.
-
Key words: Blastocystis, alpaca (Vicugna pacos), prevalence, subtype, Shanxi
Blastocystis, an enteric protist, is commonly found in humans and many animal species [
1–
3]. Although
Blastocystis can be found in asymptomatic individuals, a number of studies have shown that
Blastocystis can cause gastrointestinal symptoms, such as constipation, diarrhea, flatulence, and abdominal pain [
4–
6]. Extraintestinal manifestations such as chronic urticaria may be also associated with the presence of
Blastocystis [
6].
Blastocystis was listed in the Water, Sanitation, and Health programs of the World Health Organization in 2008 [
7,
8].
So far, different morphological forms of
Blastocystis exist, such as vacuolated (VF), granular (GF), amoebic (AF), and cystic (CF) forms have been reported [
9]. Of these forms, the robust cystic form is known to transmit infection, and the fecal-oral route is a major mode of transmission [
9,
10].
Polymerase chain reaction (PCR)-based methods using feces or fecal culture have been developed for the diagnosis of
Blastocystis infection. Also, subtyping based on the small subunit rRNA gene (SSU-rRNA) has enabled the identification of 26 distinct
Blastocystis subtypes (STs) in humans and animals [
11–
13]. Ten subtypes (ST1 to ST9 and ST12) have been identified in humans [
14,
15]. Some of these subtypes have been also reported in animals, such as ST3, ST5, ST7, and ST8, suggesting their zoonotic potential [
16,
17].
The llama (
Lama glama) and alpaca (
Vicugna pacos) are 2 domestic species of South American camelids [
18].
Blastocystis was isolated for the first time from a llama in 1993 [
19]. However, limited information is available regarding the prevalence and genetic diversity of
Blastocystis in the New World camelids. Therefore, this study was performed to determine the prevalence and genotypes of
Blastocystis in alpacas in Shanxi Province, northern China.
During August to October 2018, a total of 366 fresh fecal samples were collected from 2 counties (305 from Yangqu county and 61 from Dai county) in Shanxi Province. Each fecal specimen was obtained individually from each animal. This work was approved by the Animal Administration and Ethics Committee of Lanzhou Veterinary Institute, Chinese Academy of Agricultural Sciences (Approval No. LVRIAEC- 2018–006). The sampled animals were handled humanely in strict accordance with the requirements of the Animal Ethics Procedures and Guidelines of the People’s Republic of China.
All collected fecal specimens were kept in sterile centrifuge tubes containing 2.5% potassium dichromate. Information about the sampling location, sampling date and sex of the animals have been also gathered. Genomic DNA isolation was performed using the commercial E.Z.N.A.® Stool DNA Kit (Omega, Bio-tek Inc., Norcross, Georgia, USA) as per the manufacturer’s specifications. DNA extracted from each sample was stored at −20°C until used for nucleic acid amplification.
PCR method based on the amplification of approximately 600 bp fragment within the small subunit ribosomal RNA (SSU rRNA) of Blastocystis was carried out using primers BLF (5′-ATCTGGTTGATCCTGCCAGT-3′) and BLR (5′-GAGCTTTTTAACTGCAACAACG-3′). PCR mix (25 μl) included 10×PCR buffer (Mg2+ free), 0.2 mM of each dNTP, 2 mM of MgCl2, 1.25 U of rTaq DNA polymerase, 2 μl of DNA template and 0.4 μM of each primer. Each PCR run included a positive control and a negative control. The amplification program consisted of a pre-heating denaturation step (94°C for 5 min), followed by 30 cycles of denaturation (94°C for 45 sec), annealing (57°C for 45 sec) and extension (72°C for 1 min). A final elongation step was carried out at 72°C for 10 min. The PCR products were checked by 2% agarose gel containing ethidium bromide and visualized with UV light. All the PCR amplicons with the expected size were purified and submitted to Sangon Biotech Co. Ltd (Shanghai, China) for sequencing.
Nucleotide sequences obtained in this study were subjected to BLAST searches (
https://blast.ncbi.nlm.nih.gov/Blast.cgi). The software Clustal×1.83 was used to align the generated DNA consensus sequences with reference sequences of known
Blastocystis subtypes downloaded from GenBank to determine
Blastocystis subtypes. Phylogenetic analysis was carried out using Bayesian inference. The representative sequences in this study have been submitted to GenBank under the following accession numbers: MN382282-MN382284, MN382334-MN382337, and MN382339. Differences in infection rates in different regions and gender groups were analyzed by Chi-square (χ
2) test with the software SPSS 20.0 (IBM, Chicago, Illinois, USA), and the
P-value<0.05 was considered statistically significant.
Our data provided the first report on the presence of
Blastocystis in fecal samples of alpacas in Shanxi Province, northern China. Out of the 366 fecal samples tested, 87 (23.8%, 95% confidence interval [CI]: 19.4–28.1) were tested PCR-positive for
Blastocystis (
Table 1).
Blastocystis was detected in samples from the 2 investigated counties. The
Blastocystis prevalence in Dai county (14.8%, 95% CI: 5.9–23.7) was lower than that in Yangqu county (25.6%, 95% CI: 20.7–30.5), but the difference was not statistically significant (
P>0.05). It is difficult to explain the reasons resulting in this difference, it may be related to different stocking density, usage of medication and contact with contagium. However, there was significant difference (
P<0.01) in the prevalence of
Blastocystis between male 33.7% (95% CI: 24.0–43.4) and female 20.4% (95% CI: 15.7–25.2) alpacas. Gender-related differences in the subtype distribution of
Blastocystis have been also previously reported. For example, ST3 was found to be more common in men than in women in Sweden [
20]. Given the significant difference in
Blastocystis prevalence between male and female alpacas, we were also interested in testing whether there were any gender differences in the occurrence of ST5, ST10, and ST14. However, no statistically significant difference in the prevalence of these STs were detected.
The prevalence of
Blastocystis in alpacas from Shanxi Province (23.8%) was lower than that reported in alpacas (85.7%), camels (50.0%), and guanacos (70.0%) in Shaanxi Province, but was similar to that in camels in Libya (24.0%) [
21,
22]. Differences in the prevalence of
Blastocystis among studies may be attributed to sample numbers since in the study of Zhao et al. included only 14 alpacas [
22], whereas in the present study fecal samples were examined from 366 alpacas. The differences in prevalence may also result from variations in the geographical locations, management approaches, climatic conditions, sampling seasons, and the type of the camelids. Further epidemiological investigations of
Blastocystis in alpacas and other camelid species should be performed in order to determine the reasons for differences in
Blastocystis prevalence.
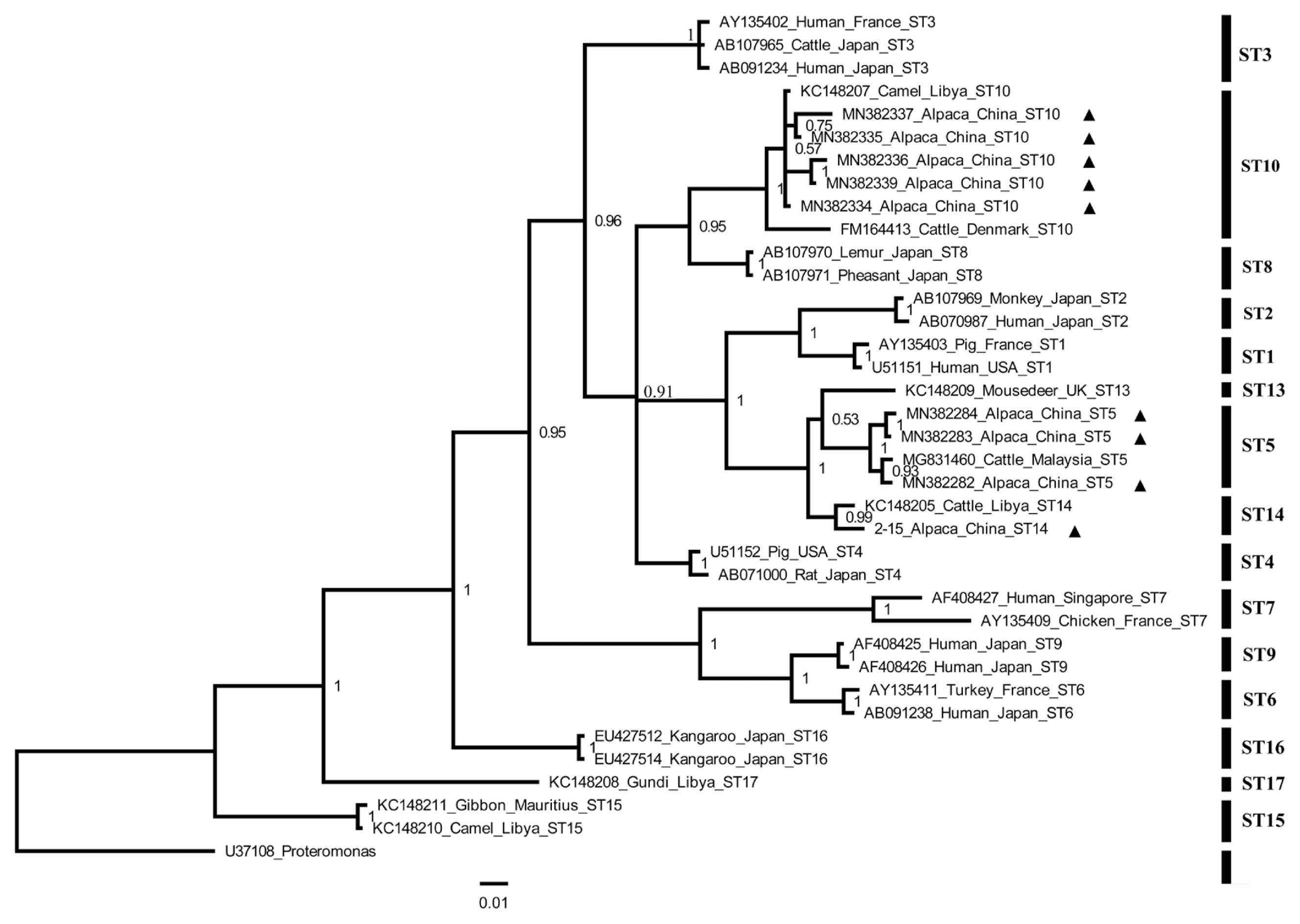
BLAST analysis identified 3
Blastocystis subtypes (namely ST5, ST10, and ST14) from alpacas in Shanxi Province. This finding was confirmed by phylogenetic analysis (
Fig. 1). The ST10 was the most dominant subtype found in animals belonging to artiodactyla, such as sheep in the UK and cattle from China, UK, Libya, and Denmark [
21,
23,
24]. This fact was in agreement with our results where the ST10 was the dominant subtype and ST14 was the second highly frequent subtype. These results are also consistent with the results of previous study in alpacas from Qinling Wildlife Park in Shaanxi [
22]. Interestingly, this is the first study to report the presence of
Blastocystis ST5 in alpacas. Animals have been suggested as a reservoir for transmission of
Blastocystis to humans largely because people can harbor identical or similar STs to the in-contact animals [
16,
25]. Given the zoonotic potential of ST5 [
16,
25], our results provide a new clue for the possible role of alpacas harboring
Blastocystis ST5 in the transmission of infection to humans.
In conclusion, this study revealed 23.8% Blastocystis prevalence in alpacas in Shanxi Province, northern China. We also observed gender differences in Blastocystis prevalence. Three Blastocystis subtypes (namely ST5, ST10, and ST14) were identified, and ST10 was the most dominant subtype. The fact that ST5 is a potential zoonotic subtype provides support for the potential role of alpacas harboring ST5 to serve as a source for human infection with Blastocystis. These findings extend the current knowledge of the prevalence and subtype distribution of Blastocystis in alpacas. Further studies are warranted to investigate the genetic diversity of Blastocystis in alpacas and to elucidate the role of alpacas in zoonotic transmission of Blastocystis by examining samples from alpacas and their in-contact humans from other geographical region.
Notes
-
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ACKNOWLEDGMENTS
Project support was provided, in part, by the Agricultural Science and Technology Innovation Program (ASTIP) (Grant No. CAAS-ASTIP-2016-LVRI-03), the Science and Technology Key Research and Development Project of Shanxi Province (201703D221024-2), and the Program for the Science and Technology Innovation of Shanxi Agricultural University (2017YJ10). The authors thank Associate Professor Hany M. Elsheikha in the Faculty of Medicine and Health Sciences, School of Veterinary Medicine and Science, University of Nottingham, UK, for improving the English of the revised manuscript.
Fig. 1Phylogenetic tree of Blastocystis constructed using Bayesian inference based on Blastocystis DNA sequences obtained in the present study (marked with black triangle) and sequences obtained from the NCBI. Proteromonas (U37108) was used as outgroup. Number on branch represents a support value.
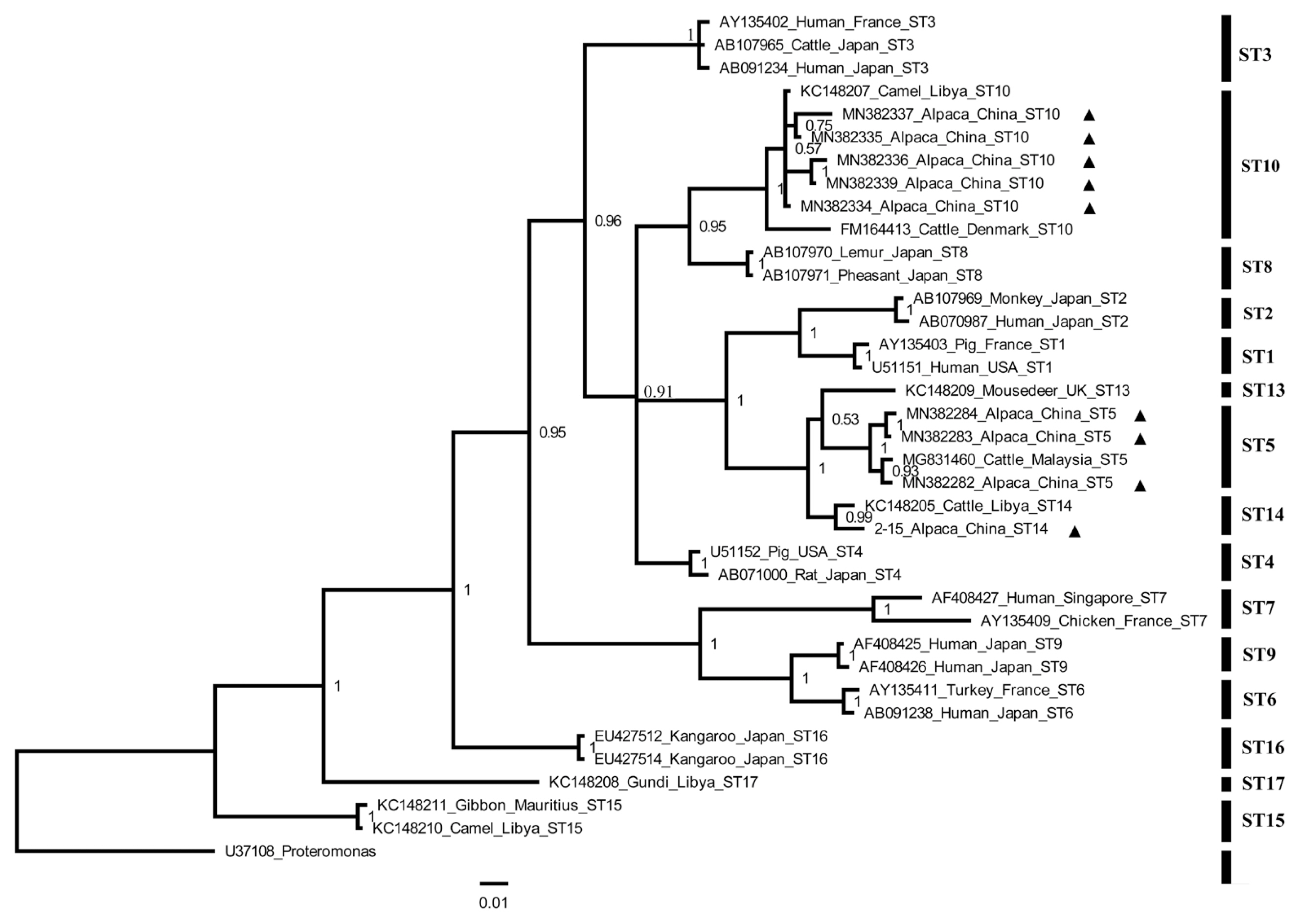
Table 1Variables associated with the prevalence and subtypes of Blastocystis in alpacas in Shanxi Province, northern China
Table 1
|
Factor |
Category |
No. of tested |
No. of positive |
Prevalence % (95% CI) |
OR (95% CI)*
|
P-value |
Subtypes (No.) |
|
Region |
Yangqu County |
305 |
78 |
25.6 (20.7–30.5) |
2.00 (0.9–4.2) |
0.07 |
ST5 (12); ST10 (37); ST14 (29) |
|
Dai County |
61 |
9 |
14.8 (5.9–23.7) |
Reference |
|
ST10 (2); ST14 (7) |
|
|
Gender |
Male |
92 |
31 |
33.7 (24.0–43.4) |
1.98 (1.2–3.3) |
<0.01 |
ST5 (3); ST10 (17); ST14 (11) |
|
Female |
274 |
56 |
20.4 (15.7–25.2) |
Reference |
|
ST5 (9); ST10 (22); ST14 (25) |
|
|
Total |
|
366 |
87 |
23.8 (19.4–28.1) |
|
|
ST5 (12); ST10 (39); ST14 (36) |
References
- 1. Amin OM. Seasonal prevalence of intestinal parasites in the United States during 2000. Am J Trop Med Hyg 2002;66:799-803.
- 2. Abe N. Molecular and phylogenetic analysis of Blastocystis isolates from various hosts. Vet Parasitol 2004;120:235-242.
- 3. Parkar U, Traub RJ, Vitali S, Elliot A, Levecke B, Robertson I, Geurden T, Steele J, Drake B, Thompson RC. Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet Parasitol 2010;169:8-17.
- 4. Boreham PF, Stenzel DJ.
Blastocystis in humans and animals: morphology, biology, and epizootiology. Adv Parasitol 1993;32:1-70.
- 5. Yakoob J, Jafri W, Beg MA, Abbas Z, Naz S, Islam M, Khan R. Irritable bowel syndrome: is it associated with genotypes of Blastocystis hominis
. Parasitol Res 2010;106:1033-1038.
- 6. Casero RD, Mongi F, Sánchez A, Ramírez JD.
Blastocystis and urticaria: examination of subtypes and morphotypes in an unusual clinical manifestation. Acta Trop 2015;148:156-161.
- 7. Poirier P, Wawrzyniak I, Vivarès CP, Delbac F, El Alaoui H. New insights into Blastocystis spp.: a potential link with irritable bowel syndrome. PLoS Pathog 2012;8:e1002545.
- 8. Deng L, Chai Y, Zhou Z, Liu H, Zhong Z, Hu Y, Fu H, Yue C, Peng G. Epidemiology of Blastocystis sp. infection in China: a systematic review. Parasite 2019;26:41.
- 9. Nagel R, Gray C, Bielefeldt-Ohmann H, Traub RJ. Features of Blastocystis spp. in xenic culture revealed by deconvolutional microscopy. Parasitol Res 2015;114:3237-3245.
- 10. Palasuwan A, Palasuwan D, Mahittikorn A, Chiabchalard R, Combes V, Popruk S. Subtype Distribution of Blastocystis in Communities along the Chao Phraya River, Thailand. Korean J Parasitol 2016;54:455-460.
- 11. Roberts T, Stark D, Harkness J, Ellis J. Subtype distribution of Blastocystis isolates from a variety of animals from New South Wales, Australia. Vet Parasitol 2013;196:85-89.
- 12. Li WC, Wang K, Gu Y. Occurrence of Blastocystis sp. and Pentatrichomonas hominis in sheep and goats in China. Parasit Vectors 2018;11:93.
- 13. Maloney JG, Lombard JE, Urie NJ, Shivley CB, Santin M. Zoonotic and genetically diverse subtypes of Blastocystis in US pre-weaned dairy heifer calves. Parasitol Res 2019;118:575-582.
- 14. Ramírez JD, Sánchez A, Hernández C, Flórez C, Bernal MC, Giraldo JC. Geographic distribution of human Blastocystis subtypes in South America. Infect Genet Evol 2016;41:32-35.
- 15. Zhang W, Ren G, Zhao W, Yang Z, Shen Y, Sun Y, Liu A, Cao J. Genotyping of Enterocytozoon bieneusi and subtyping of Blastocystis in cancer patients: relationship to diarrhea and assessment of zoonotic transmission. Front Microbiol 2017;8:1835.
- 16. Yan Y, Su S, Ye J, Lai X, Lai R, Liao H, Chen G, Zhang R, Hou Z, Luo X.
Blastocystis sp. subtype 5: a possibly zoonotic genotype. Parasitol Res 2007;101:1527-1532.
- 17. Moosavi A, Haghighi A, Mojarad EN, Zayeri F, Alebouyeh M, Khazan H, Kazemi B, Zali MR. Genetic variability of Blastocystis sp. isolated from symptomatic and asymptomatic individuals in Iran. Parasitol Res 2012;111:2311-2315.
- 18. Kadwell M, Fernandez M, Stanley HF, Baldi R, Wheeler JC, Rosadio R, Bruford MW. Genetic analysis reveals the wild ancestors of the llama and the alpaca. Proc Biol Sci 2001;268:2575-2584.
- 19. Stenzel DJ, Cassidy MF, Boreham PF. Morphology of Blastocystis sp. isolated from circus animals. Int J Parasitol 1993;23:685-687.
- 20. Forsell J, Granlund M, Stensvold CR, Clark CG, Evengård B. Subtype analysis of Blastocystis isolates in Swedish patients. Eur J Clin Microbiol Infect Dis 2012;31:1689-1696.
- 21. Alfellani MA, Taner-Mulla D, Jacob AS, Imeede CA, Yoshikawa H, Stensvold CR, Clark CG. Genetic diversity of Blastocystis in livestock and zoo animals. Protist 2013;164:497-509.
- 22. Zhao GH, Hu XF, Liu TL, Hu RS, Yu ZQ, Yang WB, Wu YL, Yu SK, Song JK. Molecular characterization of Blastocystis sp. in captive wild animals in Qinling Mountains. Parasitol Res 2017;116:2327-2333.
- 23. Stensvold CR, Alfellani MA, Nørskov-Lauritsen S, Prip K, Victory EL, Maddox C, Nielsen HV, Clark CG. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int J Parasitol 2009;39:473-479.
- 24. Zhu W, Tao W, Gong B, Yang H, Li Y, Song M, Lu Y, Li W. First report of Blastocystis infections in cattle in China. Vet Parasitol 2017;246:38-42.
- 25. Wang W, Owen H, Traub RJ, Cuttell L, Inpankaew T, Bielefeldt-Ohmann H. Molecular epidemiology of Blastocystis in pigs and their in-contact humans in Southeast Queensland, Australia, and Cambodia. Vet Parasitol 2014;203:264-269.