Abstract
Spirometra tapeworms (Cestoda: Diphyllobothriidae) collected from carnivorous mammals in Tanzania were identified by the DNA sequence analysis of the mitochondrial cytochrome c oxidase subunit 1 (cox1) and internal transcribed spacer 1 (ITS1), and by morphological characteristics. A total of 15 adult worms were collected from stool samples and carcasses of Panthera leo, Panthera pardus, and Crocuta crocuta in the Serengeti and Selous ecosystems of Tanzania. Three Spirometra species: S. theileri, S. ranarum and S. erinaceieuropaei were identified based on morphological features. Partial cox1 sequences (400 bp) of 10 specimens were revealed. Eight specimens showed 99.5% similarity with Spirometra theileri (MK955901), 1 specimen showed 99.5% similarity with the Korean S. erinaceieuropaei and 1 specimen had 99.5% similarity with Myanmar S. ranarum. Sequence homology estimates for the ITS1 region of S. theileri were 89.8% with S. erinaceieuropaei, 82.5% with S. decipiens, and 78.3% with S. ranarum; and 94.4% homology was observed between S. decipiens and S. ranarum. Phylogenetic analyses were performed with 4 species of Spirometra and 2 species of Dibothriocephalus (=Diphyllobothrium). By both ML and BI methods, cox1 and ITS1 gave well supported, congruent trees topology of S. erinaceieuropaei and S. theileri with S. decipiens and S. ranarum forming a clade. The Dibothriocephalus species were sisters of each other and collectively forming successive outgroups. Our findings confirmed that 3 Spirometra species (S. theileri, S. ranarum, and S. erinaceieuropaei) are distributed in the Serengeti and Selous ecosystems of Tanzania.
-
Key words: Spirometra erinaceieuropaei, Spirometra ranarum, Spirometra theileri, cox1, ITS1
INTRODUCTION
Tapeworms of the genus
Spirometra (Cestoda: Diphyllobothriidae) are intestinal parasites of feline and canine mammals. They have complex life cycles involving 2 intermediate hosts, such as freshwater copepods (
Cyclops spp.), followed by amphibians (tadpoles and frogs) amphibians (tadpoles and frogs) or reptiles (snakes) that consume copepods whilst drinking water [
1]. The plerocercoid larva, the sparganum, frequently infects humans by the consumption of raw or undercooked meat from the second intermediate or transport host, or consumption of untreated water containing procercoid-infected copepods [
2].
Few species of
Spirometra have been reported in Africa.
S. pretoriensis (=
Lühella pretoriense) has been found in the bat-eared fox (
Otocyon megalotis) in South Africa (Baer, 1924, 1926) [
3,
4] and in the African wild dog (
Lycaon pictus) in DRC Congo (Baer and Fain, 1955) [
5].
S. theileri (=
Diphyllobothrium theileri) has been found in the bush cat (
Leptailurus serval) and the tiger cat (
Felis lybica) near Pretoria, and in the lion (
Panthera leo) in DRC Congo (Baer, 1959) [
6]. The plerocercoids of
Spirometra, known as spargana, have been detected in wild herbivorous mammals such as buffalo (
Syncerus caffer), zebra (
Equus quagga), wildebeest (
Connochaetes taurinus), and warthogs (
Phacochoerus africanus) in Tanzania and Kenya [
7,
8]. Adults of
S. theileri were collected from dogs and cats experimentally infected with spargana from warthogs (
Phacochoerus africanus) in Tanzania [
9].
S. pretoriensis was found in hyena in Ethiopia [
10]. The eggs and proglottids of
Spirometra had been found in fecal samples of spotted hyenas (
Crocuta crocuta) and lions in previous studies [
8,
11,
12]. A high prevalence of
Spirometra infection was reported in lions from the Serengeti and Ngorongoro Crater of Tanzania [
13].
Spirometra species was commonly found in the intestines of lion and wild dogs in the Luangwa Valley, Zambia [
14]. In lions from Tarangire, northern Tanzania, the prevalence of
Spirometra was higher than that of other intestinal helminths [
15].
Previous studies reported
S. ranarum from lions in the Seronera area [
16] and
S. theileri from leopard (
Panthera pardus) and spotted hyena in Maswa Game Reserve [
17], which were identified by molecular analysis of mitochondrial
cox1 and NADH dehydrogenase subunit 1(
nad 1), together with morphological observations. Both of these areas are within the Serengeti ecosystem, which suggested that further studies were needed to clarify the diversity of
Spirometra in the various geographical areas of Tanzania.
In this study, we investigated the occurrence of Spirometra in carnivorous mammals (the main definitive hosts for Spirometra) in 2 widely separated and ecologically important areas, the Serengeti ecosystem in northern Tanzania, and the Selous ecosystem in the far eastern part of Tanzania.
MATERIALS AND METHODS
Sample collection and morphological analysis
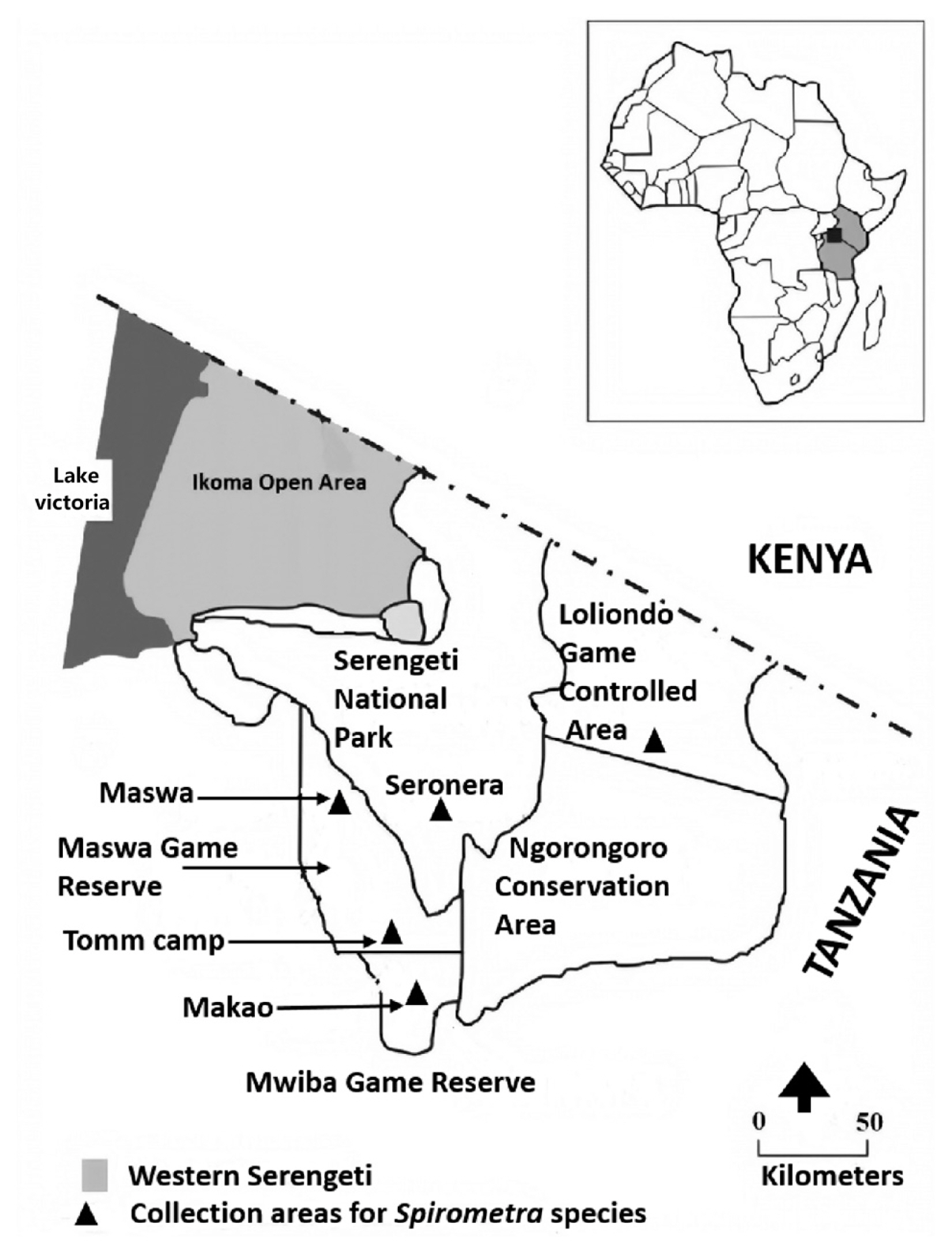
Tapeworm sample collection was carried out in the Selous and Serengeti ecosystems in Tanzania between 2011 and 2018. These areas were selected owing to its diverse wildlife species. A total of 15
Spirometra samples was collected from the stool and the intestines of the leopard,
Panthera pardus, (n=8), spotted hyena,
Crocuta crocuta, (n=1), and lion,
Panthera leo, (n=6). Of 15 samples, 12 were obtained from Loliondo, Maswa, Tomm camp, and Tototo in the Serengeti ecosystem, and 3 were obtained from Utunge Kingupira in the Selous ecosystem (
Fig. 1;
Table 1).
The collected worms were preserved separately in 70% ethanol and 10% formalin for molecular and morphological observations. For morphological observations, each gravid proglottid of 15 samples preserved in 10% formalin was gently compressed between 2 glass slides before carmine staining. The morphological characteristics of the samples were compared with those described by Baer 1926 [
4] and Faust et al. 1929 [
18] for carmine-stained specimens, including the width and length of mature and gravid proglottids, number of uterine loops, position of the uterine pore, genital pore, vaginal opening, size and number of testes, and size of eggs.
Genomic DNA was extracted individually from 15 adult tapeworms using a QIAamp DNA Mini Kit following the manufacturer’s instructions (Qiagen, Valencia, California, USA). A partial sequence of the mitochondrial
cox1 gene was employed, as described by Jeon et al. [
20]. PCR primers for the amplification of internal transcribed spacer 1 (ITS1) were designed from conserved sequences of the end regions of 18S and 5.8S ribosomal DNA to examine the relationships between congeneric and other closely-related species. Primers used to amplify the ITS1 region were Di-ITS1F (5′-AAC AAG GTT TCC GTA GGT GA-3′) and Di-ITS1R (5′-AGC AGT CTG CGA TTC ACA TT-3′). The 25 μl reaction mixture contained 50 ng of nuclear DNA, 10 pmol of each ITS1 primer, 2.5 μl of 10×buffer, 12.25 μl of 2×buffer (MgCl2, dNTP), and 1.25 units of
Taq polymerase (TAKARA Bio, Inc., Kusatsu, Japan). Reaction conditions were 3 min at 95°C, over 35 cycles of 1 min at 95°C, 1 min at 58°C, 1 min at 72°C, and a final extension step of 10 min at 72°C. PCR products were purified and ligated into the pGEM-T vector (Promega, Madison, Wisconsin, USA), transformed into
Escherichia coli, and plated onto LB agar containing X-Gal (20 mg/ml) and ampicillin (100 μg/ml). Plasmid DNA from selected colonies was digested by
EcoRI to confirm the size of the insert. Ten positive clones from each PCR product were used for nucleotide sequence confirmation.
DNA sequences of the mitochondrial cox1 gene and ITS1 were assembled using Geneious R9.1 (Biometer, Auckland, New Zealand). The sequences were identified by comparisons with cox1 sequences of Spirometra erinaceieuropaei, S. decipiens, S. ranarum, and S. theileri, and with ITSI sequences of Diphyllobothrium latum and Diphyllobothrium nihonkaiense in the GenBank database.
A phylogenetic analysis was performed by Bayesian inference (BI) and maximum likelihood (ML) using partial mitochondrial
cox1 (390 bp) sequences of
S. erinaceieuropaei (KJ599680),
S. decipiens (KJ599679),
S. ranarum (MH298843),
D. nihonkaiense (EF420138), and
D. latum (DQ985706) and ITS1 (526 bp) sequences of
D. nihonkaiense (AB288368 and AB288369). The ML analyses of
cox1 and ITS1 were implemented in MEGA7 [
19] with the HKY+G substitution model, determined using a Modeltest. BI analyses were performed using Bayesian Evolutionary Analysis Sampling Trees (BEAST) (version 1.10.4) [
20,
21] with the HKY substitution, which was chosen using MEGA. Nodes were assessed by bootstrapping with 1,000 pseudoreplicates.
RESULTS
Morphological features of Spirometra species in Tanzania
A strobila of
S. erinaceieuropaei collected from a leopard measured 45 cm long. The scolex was spoon-shaped, measuring 0.4 mm in diameter and 1.0 mm in length. The proglottid segments measured 5 to 9 mm in width and 5 to 8 mm in length and became broad on the terminal gravid segments. The cirrus pouch appeared just behind the male genital opening close to the vaginal pore, projecting towards the vaginal duct. The uterus covered a narrow field comprising 7 to 8 complete coils, with 4 internal loose coils located slightly behind the outer pile. The eggs were numerous, ellipsoidal, operculated, and up to 57 μm with a diameter of 34 μm, and testes and vitellaria extended to the lateral margin of the uterus and united at the anterior portion (
Fig. 2A).
A strobila of
S. ranarum collected from the stool of a lion was 75 cm long and 3 mm wide. The mature and gravid proglottids were 12.0 mm in width and 3 mm in length, on average. The uterus comprised 3–3.5 loops with a butterfly or bow-tie appearance, and adopted a diagonal direction in the second turn. The genital and vaginal pores were situated ventrally on the midline in the anterior third of the proglottid. The male genital aperture extended to the anterior border of the segment and the median was close to the female aperture on its lateral side. The testes measured 74 μm in diameter. The ovaries were highly dendritic and connected to the uterus. The eggs were numerous, 57 to 60 μm in length and 34.2 to 35.3 μm in transverse diameter (
Fig. 2B;
Table 2).
The strobilae of
S. theileri collected from the intestine of a leopard and spotted hyena were 45 cm and 134 cm in length, respectively, with varying shapes and sizes. The first segment measured 0.4 mm wide and 0.02 mm long, up to 0.6 to 1.0 mm in width, and 0.15 to 0.25 mm in length. The uterus consisted of 3–4.5 loops on each side of the proglottids. The genital pore was located ventrally on the midline in the anterior 1/5 of the proglottids. The vaginal pore was behind the male genital pore. The uterine pore was situated posterior to the vagina, and numerous thick-shelled and operculated eggs were present, 54 to 59 μm in length and 20 to 24 μm in width. The dumbbell-shaped ovary was located near the posterior margin of the proglottid and connected to the uterus (
Fig. 2C;
Table 2).
Among the 15 samples, 10 partial
cox1 sequences were successfully obtained and deposited in GenBank (under accession numbers MT775875 for
S. erinaceieuropaei, MT775874 for
S. ranarum, and MT775870-775873, MT775876-775879 for
S. theileri). Partial
cox1 sequences (−400 bp) from 8 isolates from Tanzania showed 99.5% (400/430) similarity with
cox1 of previously sequenced Tanzanian
S. theileri (MK955901). Additionally,
cox1 of one specimen (No. B473) showed 99.5% similarity with that of Korean
S. erinaceieuropaei (KJ599680), and
cox1 of another specimen (No. 457-3) showed 99.5% similarity with that of Myanmar
S. ranarum (MH298843). Sequence differences in the partial
cox1 sequence of Tanzanian
Spirometra ranged from 0% to 10.1%. Samples were identified as
S. theileri (n=8),
S. erinaceieuropaei (n=1), and
S. ranarum (n=1) by
cox1 sequence analysis.
Spirometra theileri,
S. erinaceieuropaei and
S. ranarum were all found in Serengeti ecosystem while only
S. theileri was revealed in Selous ecosystem (
Table 1).
The ITS1 sequences of 4 Spirometra species, S. theileri (MN737751, 716 bp), S. erinaceieuropaei (MN733011, 732 bp), S. decipiens (MN733010, 801 bp), and S. ranarum (MN733012, 842 bp), showed length differences. The ITS1 sequence of S. theileri showed 89.8% similarity with that of S. erinaceieuropaei (MN733011), 82.5% similarity with S. decipiens (MN733010), and 78.3% similarity with S. ranarum (MN733012), and 94.4% similarity was observed between S. decipiens and S. ranarum.
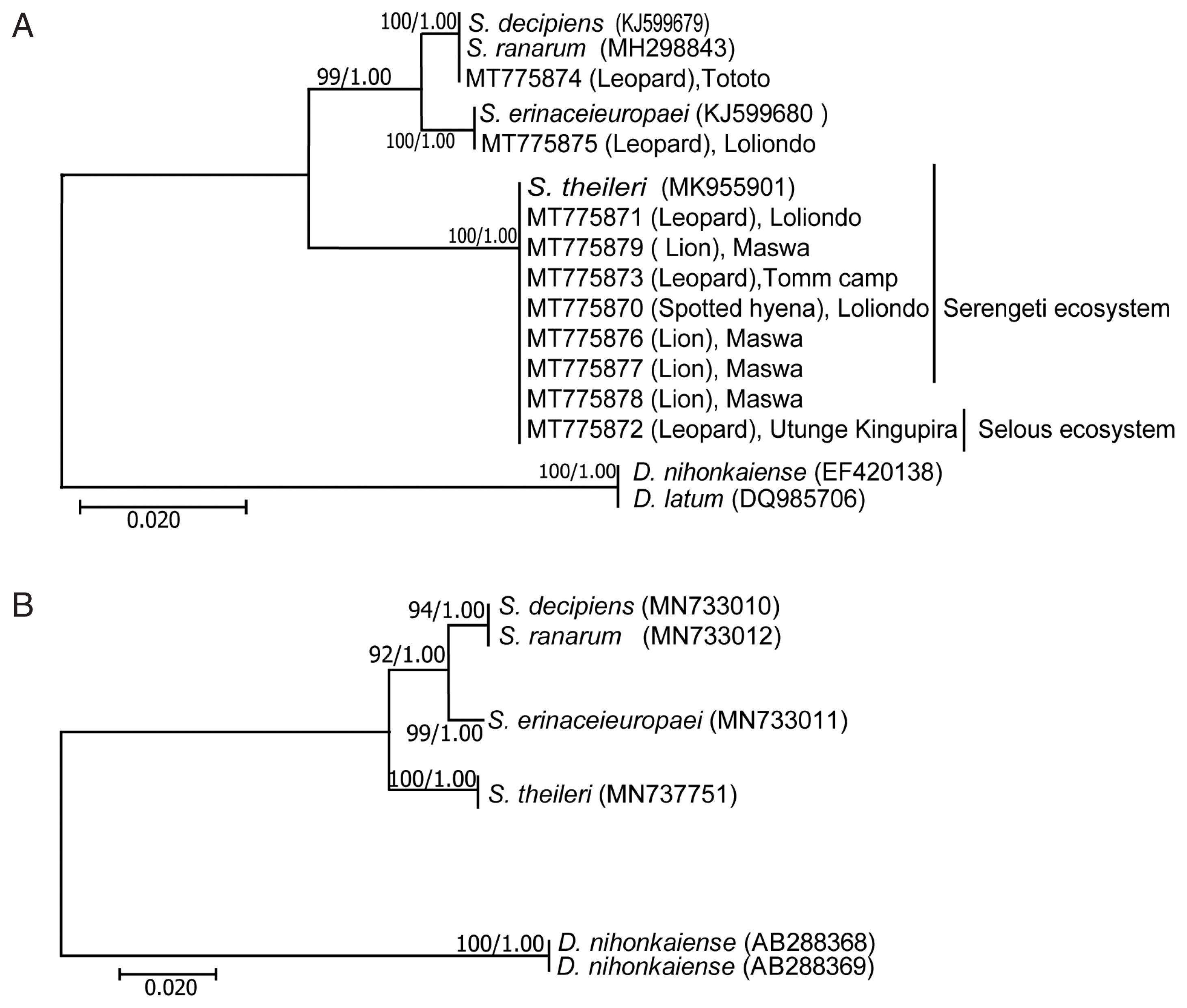
Phylogenetic analyses were performed with 4 species of Spirometra and 2 species of. Dibothriocephalus (=Diphyllobothrium).
The
cox1 and ITS1, by both ML and BI methods, gave well supported, congruent trees topology of
S. erinaceieuropaei and
S. theileri with
S. decipiens and
S. ranarum forming a clade (
Fig. 3). The
Dibothriocephalus species were sisters of each other, and, collectively, forming successive outgroups.
DISCUSSION
Spirometra can be identified based on morphological and molecular characters. In this study, S. erinaceieuropaei, S. theileri, and S. ranarum were obtained from lions, spotted hyenas, and leopards in the Selous and Serengeti ecosystems, the 2 most important ecosystems in Tanzania, with a high diversity of wildlife species. Adult Spirometra were analyzed based on genital morphology and mitochondrial cox1 and ITS1 sequences. Partial cox1 sequences obtained from 10 of the 15 samples revealed 3 species of Spirometra were present.
Based on morphological observations, uterine shapes of collected
Spirometra species vary with respect to the number of coiled shapes, with 7–8 complete coils in
S. erinaceieuropaei, 3–3.5 in
S. ranarum, and 3 on one side and 4 complete coils on another side in
S. theileri, similar to previous findings [
5,
22]. The ovaries were clearly dendritic in shape, but the eggs were operculate with slight differences in size among taxa.
Major differences between S. theileri and S. pretoriensis included the number of uterine loops and shape of the cirrus pouch. The uterus of S. theileri consisted of 3–4.5 coils and a cirrus pouch with communication through a short canal with a second smaller vesicular seminis, while the uterus of S. pretoriensis formed a single large loop on either side and the cirrus pouch contained a large vesicular seminis of variable shape. The distinct morphological features of S. ranarum relative to other Spirometra species included posterior uterine coils larger than the terminal uterine ball and coiling of the uteri diagonally rather than spirally. The widths of gravid proglottids and the uterine ball of S. ranarum were larger than those of other Spirometra species, including S. erinaceieuropaei, S. decipiens, S. theileri, and S. pretoriensis.
Mitochondrial
cox1 and
nad1 sequences of
S. ranarum and
S. theileri have been reported [
16,
17]. Our results provide a basis for the accurate molecular identification of
Spirometra species from Tanzania by providing sequences for ITS1 and partial
cox1 of 10 specimens. The 3
Spirometra species identified have now been found in Tanzania and various Asian countries, including China, Japan, Korea, Laos, and Thailand. The genetic divergence of the partial
cox1 sequences among the several species of
Spirometra (
S. erinaceieuropaei,
S. decipiens,
S. ranarum, and
S. theileri) ranged from 0% to 10.1%. Phylogenetic analyses based on mitochondrial DNA sequences with ML and BI methods showed that
S. erinaceieuropaei and
S. theileri were separated from other
Spirometra species, and
S. decipiens and
S. ranarum belonged to the same clade.
In addition to Seronera and Maswa, which have been evaluated in previous studies [
16,
17], our study is reporting 4 new endemic areas of
Spirometra species such as Loliondo Game Controlled Area, Tomm camp, and Tototo in the Serengeti ecosystem in northern Tanzania and Utunge-Kingupira in Selous ecosystem in far eastern Tanzania. Among the 15 specimens collected, 13 specimens were identified as
S. theileri collected from the lion (n=6) in Maswa area, 3 from the leopard in Utunge-Kingupira area, 2 from the leopard in Tomm camp area and 2 specimens collected from leopard (n=1) and spotted hyena (n=1) in Loliondo area.
S. ranarum was obtained from a leopard in the Tototo area and
S. erinaceieuropaei was obtained from a leopard in the Loliondo area.
In conclusion, we provide the first evidence that the leopard is a definitive host of S. erinaceieuropaei, S. theileri and S. ranarum and clarified the diversity of Spirometra species in 4 new endemic areas. These findings suggest that wild carnivorous mammals could be as reservoir hosts for Spirometra, which cause human sparganosis in Africa. The infection of wildlife species with Spirometra is likely related to the distribution of Spirometra species in various geographical locations. However, further studies are needed to evaluate the Spirometra species responsible for human sparganosis in the region.
Notes
-
CONFLICT OF INTEREST
The authors declare no conflict of interest related to this work.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF-2017K1A3A1A09085607). Materials were provided by the Parasite Resource Bank of Korea (PRB000720).
Fig. 1Map of the Serengeti ecosystem in Tanzania indicating the locations of 5 areas where 15 adult worms were collected in 2011–2018.
Fig. 2Mature proglottids of Spirometra species collected from carnivorous mammals in the Serengeti and Selous ecosystems of Tanzania (stained with acetocarmine). (A) S. erinaceieuropaei. (B) S. ranarum. (C) S. theileri. Characteristic genital organs, cirrus sac (CS), genital pore (GP), ovary (OV), vaginal pore (VP), and uterus (U) are designated in (A–C).
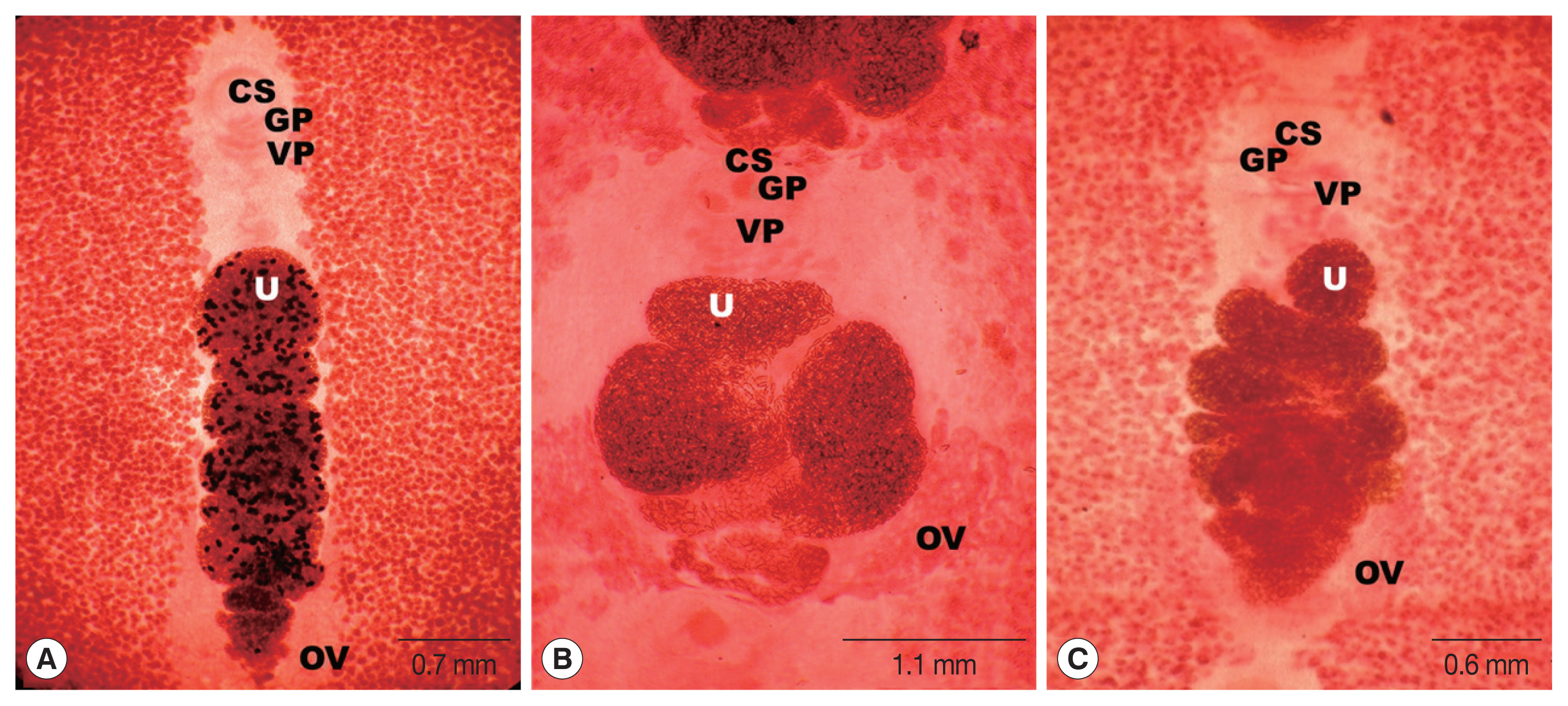
Fig. 3Phylogenetic trees of Spirometra species based on mitochondrial (A) cox1 and (B) ITS1 sequences. Numbers above the branches represent bootstrap values for trees obtained by maximum likelihood and Bayesian inference.
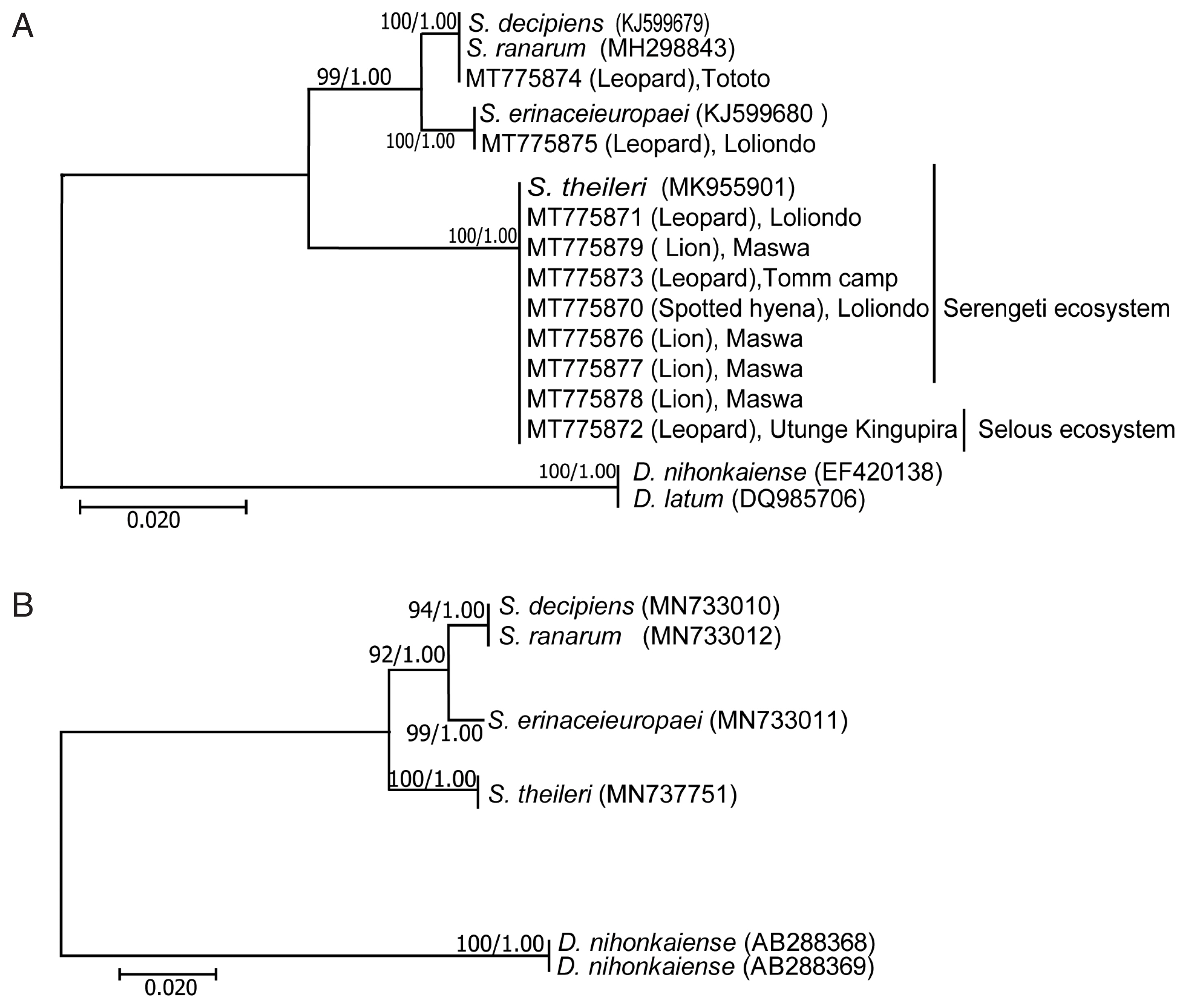
Table 1Morphological and molecular identifications of Spirometra species tapeworms by the host animals and the collecting areas in Tanzania between 2011 and 2018
Table 1
|
Host |
Collecting area |
cox1/ITS1 (GenBank No.) |
Morphology |
|
Leopard |
Loliondo |
S. erinaceieuropaei (MT775875/MN733011)b
|
S. erinaceieuropaei
|
|
Leopard |
Loliondo |
S. theileri (MT775871/MN737751)b
|
S. theileri
|
|
Spotted hyena |
Loliondo |
S. theileri (MT775870) |
S. theileri
|
|
Lion |
Maswa |
S. theileri (MT775876) |
S. theileri
|
|
Lion |
Maswa |
S. theileri (MT775877 ) |
S. theileri
|
|
Lion |
Maswa |
npa
|
S. theileri
|
|
Lion |
Maswa |
S. theileri (MT775879) |
S. theileri
|
|
Lion |
Maswa |
np |
S. theileri
|
|
Lion |
Maswa |
S. theileri (MT775878) |
S. theileri
|
|
Leopard |
Tomm camp |
S. theileri (MT775873) |
S. theileri
|
|
Leopard |
Tomm camp |
np |
S. theileri
|
|
Leopard |
Tototo |
S. ranarum (MT775874/MN733012)b
|
S. ranarum
|
|
Leopard |
Utunge, Kingupira |
S. theileri (MT775872) |
S. theileri
|
|
Leopard |
Utunge, Kingupira |
np |
S. theileri
|
|
Leopard |
Utunge, Kingupira |
np |
S. theileri
|
Table 2Morphometric features of 3 Spirometra species from carnivorous mammals in Serengeti National Park of Tanzania (unit=mm)
Table 2
|
Organs |
Organ feature |
|
S. erinaceieuropaeia
|
S. ranarumb
|
S. theileric
|
|
Scolex |
Spatulate |
Width |
0.42 |
- |
- |
|
|
Length |
1.03 |
- |
- |
|
|
Proglottids (n=10) |
Trapezoid |
Width |
5.00–9.00 |
12.00 |
0.60–0.65 |
|
|
Length |
5.00–8.00 |
3.01 |
0.15–0.25 |
|
|
Uterus |
Coiling |
Loops |
7.00–8.00 |
3.00 |
3–4.50 |
|
|
Cirrus pouch |
Oval |
Length |
|
0.17–0.25 |
0.16 |
|
|
Width |
|
0.17–0.25 |
0.13 |
|
|
Cirrus |
Cylinderical |
Length |
0.30 |
0.30 |
0.30 |
|
|
Width |
0.13 |
0.19 |
0.13 |
|
|
Seminal vesicle |
Elliptical |
Length |
0.13 |
0.19–0.22 |
0.13 |
|
|
Width |
0.14 |
0.19–0.22 |
0.14 |
|
|
Uterine terminal ball |
Oval |
Diameter |
0.06 |
0.04–0.04 |
0.07–0.08 |
|
|
Vaginal opening |
Crescentic |
Width |
0.26 |
0.25 |
0.25 |
|
|
Testes |
Polygonal |
Diameter |
0.06 |
0.06 |
0.11–0.12 |
|
|
Eggs |
Operculate |
Length |
0.06 |
0.05–0.07 |
0.05–0.06 |
|
|
Width |
0.03 |
0.03–0.04 |
0.03–0.04 |
References
- 1. Nobrega-Lee M, Hubbard G, Loverde P, Carvalho-Queiroz C, Conn DB, Rohde K, Dick EJ Jr, Nathanielsz P, Martin D, Siler-Khodr T, Schlabritz-Loutsevitch N. Sparganosis in wild-caught baboons (Papio cynocephalus anubis). J Med Primatol 2007;36:47-54. https://doi.org/10.1111/j.1600-0684.2006.00177.x
- 2. Anantaphruti MT, Nawa Y, Vanvanitchai Y. Human sparganosis in Thailand: an overview. Acta Trop 2011;118:171-176. https://doi.org/10.1016/j.actatropica.2011.03.011
- 3. Baer JG. Contribution à la faune helminthologique Sud-Africaine-Note préliminaire. Annales de Parasitologie 1924;2:237-247. (in French).
- 4. Contribution to the helminth fauna of South Africa. Mammalian cestodes. In Baer JG ed, 11th and 12th Report of the Director of Veterinary Education and Research. Pretoria, South Africa. Department of Agriculture; 1926, pp 61-136.
- 5. Baer JG, Fain A. Cestodes. Report d’Exploration Parcs Nationaux de I’Upemba. Fasc 1955;36.
- 6. Baer JG. Helminthes parasites. Exploration des Parcs Nationaux du Congo Belge. Brussels, Belgium. 1959, p 163.
- 7. Nelson GS, Pester FRN, Rickman R. The significance of wild animals in the transmission of cestodes of medical importance in Kenya. Trans R Soc Trop Med Hyg 1965;59:507-524. https://doi.org/10.1016/0035-9203(65)90153-7
- 8. Müller-Graf CD. A coprological survey of intestinal parasites of wild lions (Panthera leo) in the Serengeti and the Ngorongoro Crater, Tanzania, east Africa. J Parasitol 1995;8:812-814.
- 9. Opuni EK, Muller RL. Studies on Spirometra theileri (Baer, 1925) n. comb. 1. Identification and biology in the laboratory. J Helminthol 1974;48:15-23. https://doi.org/10.1017/s0022149x00022550
- 10. Graber M. Diphyllobothriosis and sparganosis in tropical Africa. Rev Elev Méd Vét Pays Trop 1981;34:303-311. (in French).
- 11. Bjork KE, Averbeck AG, Stromberg BE. Parasites and parasite stages of free-ranging wild lions (Panthera leo) of northern Tanzania. J Zoo Wild Med 2000;31:56-61. https://doi.org/10.1638/1042-7260(2000)031[0056:PAPSOF]2.0.CO;2
- 12. Müller-Graf CD, Woolhouse ME, Packer C. Epidemiology of an intestinal parasite (Spirometra spp.) in two populations of African lions (Panthera leo). Parasitol 1999;118:407-415. https://doi.org/10.1017/s0031182098003813
- 13. Engh AL, Nelson KG, Peebles R, Hernandez AD, Hubbard KK, Holekamp KE. Coprologic survey of parasites of spotted hyenas (Crocuta crocuta) in the Masai Mara National Reserve, Kenya. J Wildl Dis 2003;39:224-227. https://doi.org/10.7589/0090-3558-39.1.224
- 14. Berensten AR, Becker MS, Stockdale-Walden H, Matandiko W, McRobb R, Dunbar MR. Survey of gastrointestinal parasite infection in African lion (Panthera leo), African wild dog (Lycaon pictus) and spotted hyaena (Crocuta crocuta) in the Luangwa Valley, Zambia. Afr Zool 2012;47:363-368. https://doi.org/10.1080/15627020.2012.11407561
- 15. Kavana NJ, Kassuku AA, Kasanga CJ. Prevalence of Spirmetra species and other gastrointestinal helminths in wild lions (Panthera leo) in Tarangire National Park, northern Tanzania. Int J Microbiol Immunol Res 2015;4:10-14.
- 16. Eom KS, Park HS, Lee DM, Choe SJ, Kang YS, Bia MM, Lee SH, Keyyu J, Fymagwa R, Jeon HK. Molecular and morphologic identification of Spirometra ranarum found in the stool of African lion, Panthera leo in the Serengeti plain of Tanzania. Korean J Parasitol 2018;56:379-383. https://doi.org/10.3347/kjp.2018.56.4.379
- 17. Eom KS, Park H, Lee D, Choe S, Kang Y, Bia MM, Ndosi BA, Nath TC, Eamudomkarn C, Keyyu J, Fyumagwa R, Mduma S, Jeon HK. Identity of Spirometra theileri from a leopard (Panthera pardus) and spotted hyena (Crocuta crocuta) in Tanzania. Korean J Parasitol 2019;57:639-645. https://doi.org/10.3347/kjp.2019.57.6.639
- 18. Faust EC, Campbell HE, Kellogg CR. Morphological and biological studies on the species of Diphyllobothrium in China. Am J Epidemiol 1929;9:560-583. https://doi.org/10.1093/oxfordjournals.aje.a121670
- 19. Jeon HK, Park HS, Lee DM, Choe SJ, Kim KH, Huh S, Sohn WM, Chai JY, Eom KS. Human infections with Spirometra decipiens plerocercoids identified by morphologic and genetic analyses in Korea. Korean J Parasitol 2015;53:299-305. https://doi.org/10.3347/kjp.2015.53.3.299
- 20. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA7 molecular evolutionary genetics analysis using maximum likelihood evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;25:2731-2739. https://doi.org/10.1093/molbev/msr121
- 21. Suchard MA, Kitchen CMR, Sinsheimer JS, Weiss RE. Hierarchical phylogenetic models for analyzing multipartite sequence data. Systema Biol 2003;52:649-664. https://doi.org/10.1080/10635150390238879
- 22. Jeon HK, Park HS, Lee DM, Choe SJ, Kang YS, Bia MM, Lee SH, Sohn WM, Hong SJ, Chai JY, Eom KS. Genetic and morphologic identification of Spirometra ranarum in Myanmar. Korean J Parasitol 2018;56:275-280. https://doi.org/10.3347/kjp.2018.56.3.275
 , Yeseul Kang1, Tilak Chandra Nath1,3
, Yeseul Kang1, Tilak Chandra Nath1,3 , Mohammed Mebarek Bia1, Chatanun Eamudomkarn4, Hyeong-Kyu Jeon1,*
, Mohammed Mebarek Bia1, Chatanun Eamudomkarn4, Hyeong-Kyu Jeon1,* , Keeseon S. Eom1,*
, Keeseon S. Eom1,*