Abstract
Haemonchosis remains a significant problem in small ruminants. In this study, the assay of recombinase polymerase amplification (RPA) combined with the lateral flow strip (LFS-RPA) was established for the rapid detection of Haemonchus contortus in goat feces. The assay used primers and a probe targeting a specific sequence in the ITS-2 gene. We compared the performance of the LFS-RPA assay to a PCR assay. The LFS-RPA had a detection limit of 10 fg DNA, which was 10 times less compared to the lowest detection limit obtained by PCR. Out of 24 goat fecal samples, LFS-RPA assay detected H. contortus DNA with 95.8% sensitivity, compared to PCR, 79.1% sensitivity. LFS-RPA assay did not detect DNA from other related helminth species and demonstrated an adequate tolerance to inhibitors present in the goat feces. Taken together, our results suggest that LFS-RPA assay had a high diagnostic accuracy for the rapid detection of H. contortus and merits further evaluation.
-
Key words: Haemonchus contortus, goat, recombinase polymerase amplification, lateral flow strip, rapid detection
Haemonchus contortus is a blood-sucking trichostrongylid nematode parasite that infect small ruminants, leading to severe anemia, diarrhea, emaciation or even death in acute infection if left untreated [
1]. Haemonchosis causes significant losses to sheep and goat farming industry in temperate, subtropical and tropical regions, where warm and moist environment favor the survival of the free-living stages on pasture [
2].
H. contortus has a direct life-cycle that does not require an intermediate host and fertilized females can produce 5,000–7,000 eggs per day [
3]. Eggs produced by female
H. contortus are excreted in feces into the environment, where larvae hatch and develop into infective third-stage larvae (L3) within ~1 week under suitable climatic conditions (e.g., temperature and humidity). L3s of
H. contortus have a great potential to survive adverse environmental conditions such as cold or dry weather by reducing their metabolic activity and developing into a dormant state [
4,
5].
The high reproductive capacity of
H. contortus can lead to a widespread infection within and between flocks [
2]. Currently, there are 3 main classes of anthelmintic drugs (i.e. benzimidazoles, imidazothiazoles and macrocyclic lactones) available for use against
H. contortus [
2]. Unfortunately,
H. contortus has developed ability to resist all 3 main classes of these broad-spectrum anthelmintics and reports of reduced susceptibility towards the more recently developed drug class of amino-acetonitrile derivatives has also been reported [
6]. The growing rise and spread of anthelmintic resistance (AR) necessitates the development of effective diagnostic tools to rapidly and accurately identify individual animals infected by
H. contortus. The availability of such parasite detection methods could improve parasite management, reduce overuse of anthelmintics, which has driven AR, and improve disease burden estimates and surveillance in farms and areas where
H. contortus is endemic, so that informed decisions can be made surrounding the strategic use of anthelmintics.
Traditional diagnostic methods rely on microscopic identification of eggs and L3, which requires some training and have a long turnaround time [
2,
7]. To improve the accuracy of
H. contortus detection, molecular techniques based on conventional polymerase chain reaction (PCR) have been developed [
8]. PCR assays provide sensitive and specific results in a shorter time, however, PCR requires a substantial laboratory capacity, which limit the applications of PCR-based analysis in clinical settings. Loop-mediated isothermal amplification (LAMP), a novel DNA amplification method with good sensitivity and specificity was developed for detection of
H. contortus [
1,
9]. LAMP assay can be performed under isothermal conditions and the amplified products can be visualized by the naked eye [
10]. However, the primer design of LAMB assay is cumbersome, and the amplification reaction requires more than one hour to complete.
Recombinase polymerase amplification (RPA) is a novel isothermal amplification technology that was developed in 2006 [
11]. The amplification reaction of RPA can be performed under constant temperature between 37°C to 42°C in 20 min using simple equipment [
12]. The amplified products can be detected by agarose gel electrophoresis (AGE), using a pint-sized real-time fluorescence detection platform or visualized by a lateral flow strip (LFS) [
13–
15]. Given the need for a rapid diagnostic method for haemonchosis and the promising potential of the RPA technology, the present study was performed to develop for the first time, a RPA assay combined with LF strips (LFS-RPA) that targets the ITS-2 gene for rapid and accurate detection of
H. contortus in goat feces.
A series of LFS-RPA probes were designed based on the ITS-2 gene of
H. contortus (GenBank accession number MF398432.1) and their biophysical properties and dimer formation were evaluated by Oligoanalyzer 3.1 (IDT, Leuven, Belgium). The probe NZP (FAM5′-TATTGAGATTGACTTAGATAGTGACTTGTA-THF-GGCGACGATGTTCTT-3′-Spacer C3) used in the present study provided the best specificity and sensitivity. The forward primer used in the LFS-RPA amplification was the species-specific PCR primer HAE (5′-CAAATGGCATTTGTCTTTTAG-3′) of
H. contortus [
8]. Also, the reverse primer NZR (biotin-5′-TTAGTTTCTTTTCCTCCGCTAAATGATATG-3′) was designed based on the NC2 primer [
8], plus 10 bases from the 3′ end.
The amplification reaction of LFS-RPA was performed as described previously [
15]. Briefly, each amplification reaction was performed in a tube containing freeze-dried enzyme pellet provided in the TwistAmp
® nfo kit (TwistDx, Cambridge, UK) in a 50 μl final reaction volume containing 2.1 μl HAE (10 μM), 2.1 μl NZR (10 μM), 0.2 μl NZP (10 μM), 11.6 μl ddH2O, 29.5 μl rehydration buffer, 2.5 μl 280 mM magnesium acetate, and 2 μl DNA template. The tubes were shaken for a few seconds and then incubated at 37°C with constant shaking at 200 rpm. After 20 min, detection of the amplified products was performed using the PCRD Nucleic Acid Detector Assay kit (TwistDx). One μl of the amplification product was mixed with 79 μl of the PCRD extraction buffer. Then, the 80 μl mixture was added to the sample port of the PCRD device. After 10 mins, positive detection of
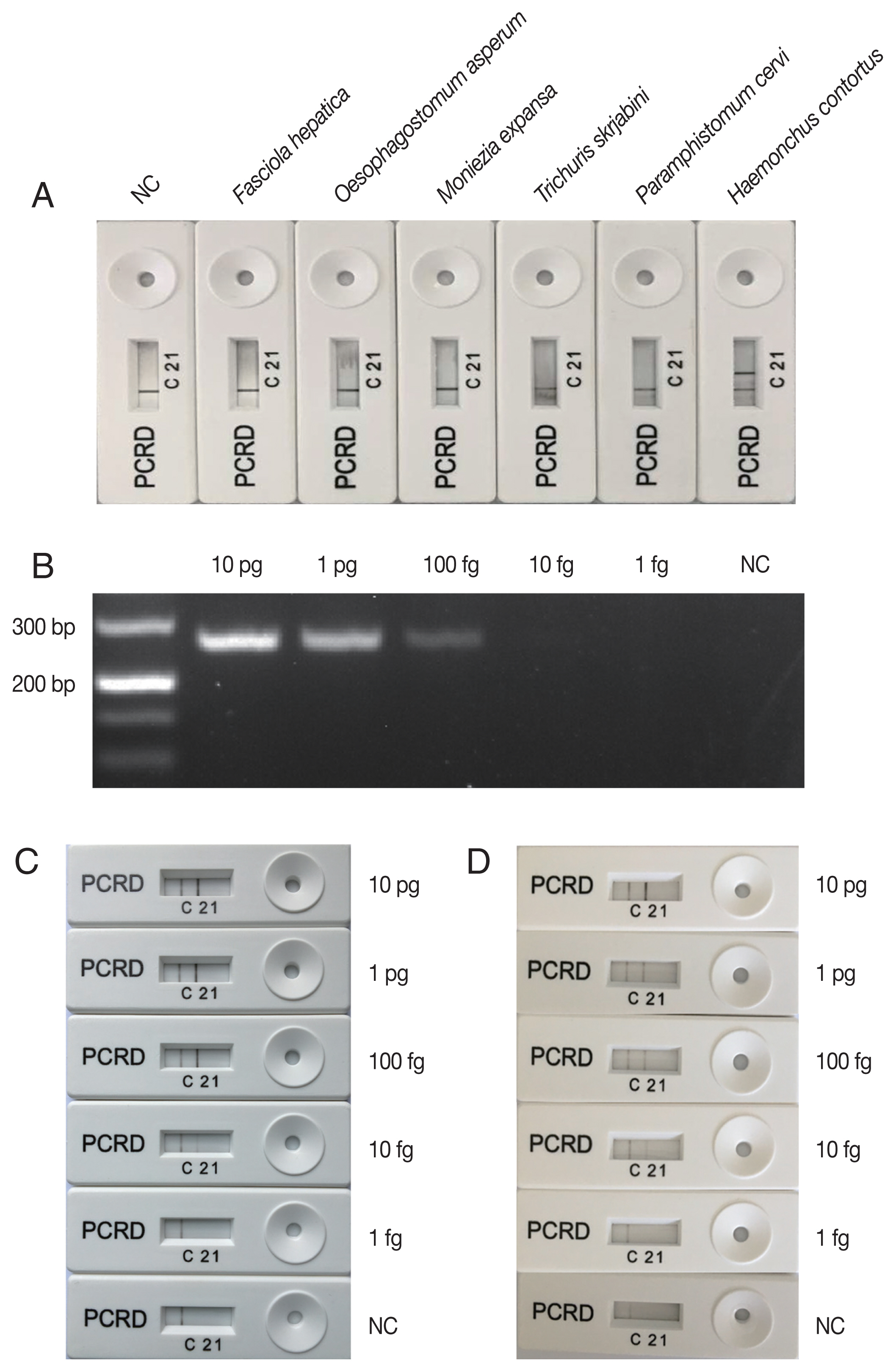
H. contortus was indicated on the PCRD device by the presence of a test line ‘2’. An additional control line ‘C’ was used to confirm that the strip assay has worked successfully. The negative result was indicated by the presence of control line ‘C’ only (
Fig. 1). A negative control sample included water only without any DNA.
Adult
H. contortus and 5 other helminth species [2 nematodes (
Oesophagostomum asperum,
Trichuris skrjabini), 2 trematodes (
Paramphistomum cervi,
Fasciola hepatica), and 1 cestode (
Moniezia expansa)] known to infect goats were collected from Tianmen city, Hubei province, China. The genomic DNAs of all the 6 helminth species were obtained by using Soil DNA kit (OMEGA, Norcross, Georgia, USA) according to the manufacturer’s recommendations, and were used to test the specificity of the LFS-RPA assay for
H. contortus. The results show that positive LFS-RPA assay was only obtained for
H. contortus DNA (
Fig. 1A), indicating that LFS-RPA primers and probe were specific to
H. contortus. However, future studies will need to include rigorous assessment of assay performance for more related trichostrongylid nematode parasites that also inhabit the ovine and caprine gut, such as species of
Trichostrongylus,
Teladorsagia,
Nematodirus,
Chabertia, and
Cooperia; and different stages of the parasite (i.e. larvae, eggs).
Sensitivity of LFS-RPA assay compared to that of a PCR was evaluated on a graded series of DNA amounts (1 fg, 10 fg, 100 fg, 1 pg, and 10 pg) in each reaction. Three reactions were performed for each DNA dilution in each assay.
H. contortus species-specific PCR conditions and primers refer to the previous study [
8]. The lowest detection level of the PCR assay was 100 fg genomic DNA (
Fig. 1B), however, visible
H. contortus-positive bands were observed on the PCRD device corresponding to LFS-RPA products amplified from 10 fg genomic DNA (
Fig. 1C), indicating that the sensitivity of LFS-RPA was about 10× higher than that of PCR, but was similar as that obtained by a LAMP assay [
1]. However, LAMP assay requires more time, a higher temperature, and more elaborate primer designing. Previous studies also showed that LFS-RPA had a more sensitivity than PCR or RT-PCR [
12,
13,
15,
16]. Interestingly,
H. contortus DNA spiked into goat feces also yielded a similar detection level (
Fig. 1D). Chemical substances found in feces, such as haeme, humic acids and bile salts, can inhibits PCR [
17,
18]. However, the results of detection of
H. contortus DNA spiked in goat feces showed that inhibitors in goat feces did not affect the detection limit of the LFS-RPA, showing the high tolerance of the assay to fecal inhibitors [
19]. LFS-RPA assay has been used to detect DNA from various sample types, such as cattle feces, human urine and environmental samples, and amplified target DNA in the presence of heparin, serum, alcohol, and haemoglobin [
12,
15,
20,
21].
The temperature and time of LFS-RPA amplification are 2 crucial parameters that we optimized in order to validate the efficiency of the assay, particularly for field applications. Different temperatures ranging from 20 to 50°C were used to determine the optimal amplification temperature. Our results showed that after amplification for 20 min, the LFS-RPA assay can still work well within a wide range of temperatures from 25°C to 45°C (
Supplementary Fig. 1A). Unlike PCR that involves amplification reaction in a time-controlled thermal gradient, the RPA worked efficiently at a wide range of temperatures, indicating that the assay can be performed without the need for heating equipment [
16,
22]. After identifying the optimal temperature for the amplification reaction, the reaction was performed for 1, 5, 10, 15, and 20 min at 39°C to determine the optimal duration of the reaction time. The amplification products can be visually detected on the PCRD device within 10 min of amplification reaction (
Supplementary Fig. 1B).
We also collected fecal samples (n=24) from goats with suspected natural H. contortus infection from Tianmen city, Hubei province, China. Total genomic DNA was extracted from each fecal sample as describe above and used as a template DNA in the amplification reactions of both the LFS-RPA and PCR assays. Twenty-three out of the 24 goat fecal samples were H. contortus-positive by LFS-RPA. However, 19 samples were H. contortus-positive by PCR. All the positive PCR products were sequenced to confirm the species identify of the worms and the obtained sequences were consistent with that of the ITS-2 gene of H. contortus. In the future, LFS-RPA will need to be compared with other detection methods, such as qPCR and LAMP, on more samples to further confirm the accuracy of this method.
Length of RPA primers and probe are suggested to be 30–35 bp and 46–52 bp, respectively, which are longer than that of PCR and RT-PCR, making the primer and probe more sensitive and specifically locate the template sequences at a low concentration. Although we did not observe any false positive results in our study, long primer and probe are more likely to form primer dimer, resulting in false positive. Hence, careful primer design is required to avoid the formation of primer dimers.
Recording results on the PCRD device was achieved by diluting 1 μl of amplicon in 79 μl of PCRD extraction buffer and adding the 80 μl of the mixture to the PCRD device, which provides more convenience compared to the old version of the lateral flow strip [
15]. The high sensitivity and specificity of LFS-RPA presents this assay as a good diagnostic tool for the detection of
H. contortus in goat feces. Interestingly, a nucleic acid purification method in under 30 sec has been developed and may provide a promising means for DNA extraction from
H. contortus eggs [
23]. Combining this quick DNA extraction method with the LFS-RPA assay would make the turnaround time of the analysis incredible short.
In conclusion, we developed a prototype LFS-RPA assay for the rapid and specific detection of H. contortus DNA in goat feces. This assay is highly sensitive, specific, with a very short turnaround time and does not require expensive equipment for the amplification reaction and detection of the amplicons. A limitation of our analysis is that the assay has been evaluated only with a set of 24 samples with unconfirmed infection status. Also, robust egg counts and larval cultures should have been performed on the fecal samples in order to get additional verification that indeed H. contortus was present in these samples. Despite these limitations, our results suggest that the LFS-RPA assay has a promising potential for rapid detection of H. contortus and warrants additional prospective analysis.
Supplementary Information
Notes
-
The authors declare that they have no competing interests.
ACKNOWLEDGMENT
This work was supported by grants from the Science and Technology plan projects of Guangdong province (2018LM2159) and the Marine Fisheries Department Guangdong projects of province (A201601A15).
Fig. 1Specificity and sensibility of LFS-RPA assay. (A) specificity of LFS-RPA assay using DNA extracted from Haemonchus contortus and 5 other taxonomically distant helminth species (Fasciola hepatica, Oesophagostomum asperum, Moniezia expansa, Trichuris skrjabini, and Paramphistomum cervi). (B) Sensitivity of the PCR compared to that of LFS-RPA assay (C) was evaluated on a series of H. contortus DNA amounts (1 fg, 10 fg, 100 fg, 1 pg, and 10 pg) in each reaction. (D) H. contortus DNA was mixed with goat faeces to test the inhibition of the LFS-RPA assay.

References
- 1. Melville L, Kenyon F, Javed S, McElarney I, Demeler J, Skuce P. Development of a loop-mediated isothermal amplification (LAMP) assay for the sensitive detection of Haemonchus contortus eggs in ovine faecal samples. Vet Parasitol 2014;206:308-312. https://doi.org/10.1016/j.vetpar.2014.10.022
- 2. Besier RB, Kahn LP, Sargison ND, Van Wyk JA. Diagnosis, treatment and management of Haemonchus contortus in small ruminants. Adv Parasitol 2016;93:181-238. https://doi.org/10.1016/bs.apar.2016.02.024
- 3. Getachew T, Dorchies P, Jacquiet P. Trends and challenges in the effective and sustainable control of Haemonchus contortus infection in sheep. Review. Parasite 2007;14:3-14. https://doi.org/10.1051/parasite/2007141003
- 4. Fakae BB. Seasonal changes and hypobiosis in Haemonchus contortus infection in the West African Dwarf sheep and goats in the Nigerian derived savanna. Vet Parasitol 1990;36:123-130. https://doi.org/10.1016/0304-4017(90)90100-P
- 5. Besier RB, Kahn LP, Sargison ND, Van Wyk JA. Chapter Four - The pathophysiology, ecology and epidemiology of Haemonchus contortus infection in small ruminants. Adv Parasitol 2016;93:95-143. https://doi.org/10.1016/bs.apar.2016.02.022
- 6. Kotze AC, Prichard RK. Chapter nine - anthelmintic resistance in Haemonchus contortus: history, mechanisms and diagnosis. Adv Parasitol 2016;93:397-428. https://doi.org/10.1016/bs.apar.2016.02.012
- 7. Demeler J, Schein E, von Samson-Himmelstjerna G. Advances in laboratory diagnosis of parasitic infections of sheep. Vet Parasitol 2012;189:52-64. https://doi.org/10.1016/j.vetpar.2012.03.032
- 8. Bott NJ, Campbell BE, Beveridge I, Chilton NB, Rees D, Hunt PW, Gasser RB. A combined microscopic-molecular method for the diagnosis of strongylid infections in sheep. Int J Parasitol 2009;39:1277-1287. https://doi.org/10.1016/j.ijpara.2009.03.002
- 9. Yang X, Qi MW, Zhang ZZ, Gao C, Wang CQ, Lei WQ, Tan L, Zhao JL, Fang R, Hu M. Development and evaluation of a loop-mediated isothermal amplification (LAMP) assay for the detection of Haemonchus contortus in goat fecal samples. J Parasitol 2017;103:161-167. https://doi.org/10.1645/16-157
- 10. Yang Y, Qin X, Zhang W, Li Z, Zhang S, Li Y, Zhang Z. Development of an isothermal recombinase polymerase amplification assay for rapid detection of pseudorabies virus. Mol Cell Probes 2017;33:32-35. https://doi.org/10.1016/j.mcp.2017.03.005
- 11. Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PloS Biol 2006;4:e204. https://doi.org/10.1371/journal.pbio.0040204
- 12. Wu YD, Zhou DH, Zhang LX, Zheng WB, Ma JG, Wang M, Zhu XQ, Xu MJ. Recombinase polymerase amplification (RPA) combined with lateral flow (LF) strip for equipment-free detection of Cryptosporidium spp. oocysts in dairy cattle feces. Parasitol Res 2016;115:3551-3555. https://doi.org/10.1007/s00436-016-5120-4
- 13. Crannell ZA, Castellanos-Gonzalez A, Irani A, Rohrman B, White AC, Richards-Kortum R. Nucleic acid test to diagnose cryptosporidiosis: lab assessment in animal and patient specimens. Anal Chem 2014;86:2565-2571. https://doi.org/10.1021/ac403750z
- 14. Teoh BT, Sam SS, Tan KK, Danlami MB, Shu MH, Johari J, Hooi PS, Brooks D, Piepenburg O, Nentwich O, Wilder-Smith A, Franco L, Tenorio A, AbuBakar S. Early detection of dengue virus by use of reverse transcription-recombinase polymerase amplification. J Clin Microbiol 2015;53:830-837. http://doi.org/10.1128/JCM.02648-14
- 15. Wu YD, Xu MJ, Wang QQ, Zhou CX, Wang M, Zhu XQ, Zhou DH. Recombinase polymerase amplification (RPA) combined with lateral flow (LF) strip for detection of Toxoplasma gondii in the environment. Vet Parasitol 2017;243:199-203. https://doi.org/10.1016/j.vetpar.2017.06.026
- 16. Gao W, Huang H, Zhu P, Yan X, Fan J, Jiang J, Xu J. Recombinase polymerase amplification combined with lateral flow dipstick for equipment-free detection of Salmonella in shellfish. Bioprocess Biosyst Eng 2018;41:603-611. https://doi.org/10.1007/s00449-018-1895-2
- 17. Wilson IG. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 1997;63:3741-3751. https://doi.org/10.1128/AEM.63.10.3741-3751.1997
- 18. Rådström P, Knutsson R, Wolffs P, Lövenklev M, Löfström C. PrePCR processing: Strategies to generate PCR-compatible samples. Mol Biotechnol 2004;26:133-146. https://doi.org/10.1385/MB:26:2:133
- 19. Katrin K, Jekaterina F, Oana T, Julia S, Taavi L, Hiljar S, Imre M, Made L, Indrek T, Ülo L. Sensitive and rapid detection of Chlamydia trachomatis by recombinase polymerase amplification directly from urine samples. J Mol Diagn 2014;16:127-135. https://doi.org/10.1016/j.jmoldx.2013.08.003
- 20. Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M. Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis. Malar J 2014;13:99. https://doi.org/10.1186/1475-2875-13-99
- 21. Rosser A, Rollinson D, Forrest M, Webster BL. Isothermal Recombinase Polymerase amplification (RPA) of Schistosoma haematobium DNA and oligochromatographic lateral flow detection. Parasit Vectors 2015;8:446. https://doi.org/10.1186/s13071-015-1055-3
- 22. Crannell ZA, Rohrman B, Richards-Kortum R. Equipment-free incubation of recombinase polymerase amplification reactions using body heat. PLoS One 2014;9:e112146. https://doi.org/10.1371/journal.pone.0112146
- 23. Zou Y, Mason MG, Wang Y, Wee E, Turni C, Blackall PJ, Trau M, Botella JR. Nucleic acid purification from plants, animals and microbes in under 30 seconds. PLoS Biol 2017;15:e2003916. https://doi.org/10.1371/journal.pbio.2003916
 , Qi-Qi Wang2, Meng Wang2, Hany M. Elsheikha3, Xin Yang4, Min Hu5, Xing-Quan Zhu2,6
, Qi-Qi Wang2, Meng Wang2, Hany M. Elsheikha3, Xin Yang4, Min Hu5, Xing-Quan Zhu2,6 , Min-Jun Xu1,*
, Min-Jun Xu1,*