Abstract
In this study, we have collected and screened a total of 268 stool samples from diarrheal patients admitted to an Infectious disease hospital in Kolkata for the presence of Cryptosporidium spp. The initial diagnosis was carried out by microscopy followed by genus specific polymerase chain reaction assays based on 70 kDa heat shock proteins (HSP70). DNA sequencing of the amplified locus has been employed for determination of genetic diversity of the local isolates. Out of 268 collected samples, 12 (4.48%) were positive for Cryptosporidium spp. Sequences analysis of 70 kDa heat shock proteins locus in 12 Cryptosporidium local isolates revealed that 2.24% and 1.86% of samples were showing 99% to 100% identity with C. parvum and C. hominis. Along with the other 2 major species one recently described globally distributed pathogenic species Cryptosporidium viatorum has been identified. The HSP70 locus sequence of the isolate showed 100% similarity with a previously described isolate of C. viatorum (Accession No. JX978274.1, JX978273.1, and JN846706.1) present in GenBank.
-
Key words: Cryptosporidium viatorum, cryptosporidiosis, molecular characterization, 70 kDa heat shock proteins (HSP70)
Cryptosporidium spp. are leading cause of diarrhoea and malnutrition, particularly in the developing countries around the world. At least 34 different species and more than 40 genotypes of
Cryptosporidium have been described to infect humans and other animals and numerous of which are reported to have zoonotic potential [
1,
2]. The majority of the infections caused by the 5 species of
Cryptosporidium are-
Cryptosporidium hominis,
Cryptosporidium parvum,
Cryptosporidium meleagridis,
Cryptosporidium felis, and
Cryptosporidium canis, of which the first 2 are the major causative agent of cryptosporidiosis in human [
3,
4].
Cryptosporidium viatorum was first described in 2012 from travellers returning to Great Britain from India, Nepal, Bangladesh and Pakistan [
5–
7].
The acute symptoms associated with cryptosporidiosis due to
C. viatorum infection in the travellers returning to Sweden and Great British from Bangladesh, India, Nepal, Pakistan Kenya, and Guatemala have included diarrhoea, abdominal pain, headache, vomiting nausea, fever and marked weight loss with illness lasting from 9 to 30 days [
6,
8]. Although it has been observed that infection with more uncommon species/genotypes is not inevitably associated with the immune status of the hosts[
9].
Cryptosporidium can be diagnosed by a number of techniques including of identification of oocysts in stools using modified acid fast staining but this technique does not allow to identify the species as the oocysts are morphologically indistinguishable and measure 5.35×4.72 μm with a length to width ratio of 1.14 [
5]. For prevention and control of cryptosporidiosis, identification and characterization of different
Cryptosporidium species along with determination of population variants (genotypes and subtypes) are fundamental in the study of cryptosporidiosis epidemiology. As the oocysts of many species of
Cryptosporidium are indistinguishable from each other, molecular techniques are necessary for identification of species and determination of genotype and subtype in order to specify the organism responsible for infection and the source and routes of transmission.
In this communication, we present a report of sequence characterization at the HSP70 locus of various Cryptosporidium isolates that have been screened by microscopy and PCR. The samples were collected from diarrheal patients admitted to an Infectious disease hospital in Kolkata. The sequence information generated from this study showed the presence of Cryptosporidium viatorum in 1 sample. Till date the presence of C. viatorum is very scarcely reported from India.
The target population of this study was patients from different parts of Kolkata and adjacent areas admitted to the Infectious Diseases Hospital (ID Hospital), Kolkata with diarrheal complaints. A total of 268 faecal samples were collected from this hospital from June, 2017 to August, 2017. This study was approved by the human Ethics Committee of I.D. Hospitalas per the Institutional Review Board (IRB Number:A-1/2015-IEC), in compliance with their ethical guidelines and in accordance with the Institutional Human Ethics Committee and written informed consent from patient has been taken also.
The parasites were detected primarily by conventional microscopy which is still considered the gold standard method for any epidemiological study although this does not allow species identification as oocysts of different Cryptosporidium species are morphologically indistinguishable. All the collected faecal samples were subjected to modified acid-fast staining and examined under an oil immersion lens of microscope in 100×magnifications. Unlike the Ziehl-Neelsen Modified Acid-Fast Stain, this stain does not require the heating of reagents for staining. After parasitological examinations all the positives samples were store in −20°C for further use. The samples that were found positive for Cryptosporidium spp. in microscopy were extracted DNA directly from the stool samples by using Stool DNA Mini Kit (QIAGEN, Valencia, California, USA) according to manufacturer’s protocol.
Further characterization of Cryptosporidium spp. was performed by PCR amplification of 70 kDa Heat Shock Protein (HSP70) followed by bi-directional sequencing of a 324 bp region of this gene. In the PCR based amplification of 324 bp of region HSP70 used genus specific primer pairs were CR_HSP70_FP 5′-GCTGGTGATACTCACTTGGG-3′ and CR_HSP70_RP5′-TCTCTTGTCCATACCAGCA-3′. PCR was carried out in a 50 μl reaction volume using 1X buffer containing 1.5 mM MgCl2 (Roche, Mannheim, Germany), 200 μM of each dNTP, 10 pmol of each primer (GCC Biotech, Joychandipur, India), 1 unit of Taq DNA polymerase enzyme (Roche) and 125 ng of isolated DNA. The PCR cycle conditions for the gene amplification were: 94°C for 3 min (initial denaturation), followed by 35 cycles of 94°C for 45 sec (denaturation), 55°C for 45 sec (annealing) and 72°C for 30 sec (extension), with a final extension of 5 min at 72°C and kept for hold at 4°C. Both positive (Cryptosporidium DNA) and negative/no template control (containing all PCR reagents without DNA template) controls have been applied in each round of PCR to validate the results. After completion of PCR, the PCR products were separated by electrophoresis for 35 min at 90 V in 1.5% agarose gel to observe specific bands in presence of marker. Amplicons representing specific band size were extracted from gels and purified by using Agarose Gel DNA Extraction Kit (Roche) according to manufacturer’s protocol and their yield were verified again by electrophoresis. The purified products were then subjected to bidirectional automated sequencing using Applied Biosystems BigDye Terminator V3.1 cycle sequencing kit (Thermo Fisher Scientific, Wiltham, Massachusetts, USA) with same primers as for PCR amplification. ABI PRISMR3100 Genetic Analyzer (Applied BioSystems,Wiltham, Massachusetts, USA) was used for the sequencing. Cryptosporidium species/genotypes were determined by comparing the resultant sequences with sequences with sequences in GenBank using BLAST database search and levels of identity established. The sequence was then aligned using MultAlign.
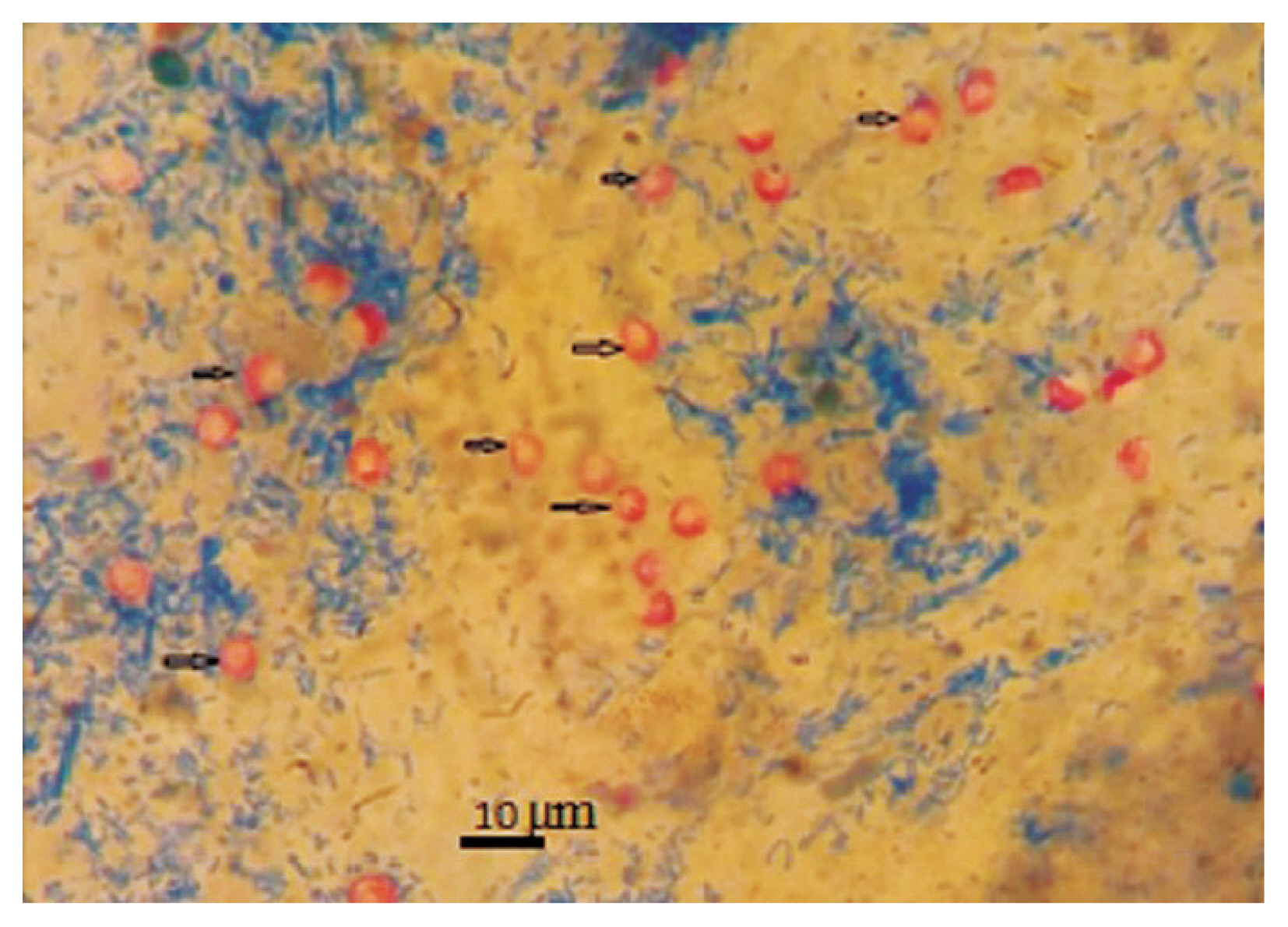
Out of 268 samples 4.5% (n/N=12/268; 95%; 95% CI 2.6% to 7.7%) were positive for
Cryptosporidium spp. by modified acid fast staining (
Fig. 1). The median age of patients in this study was 29 years (range 3–65 years). In order to consider the genetic diversity of
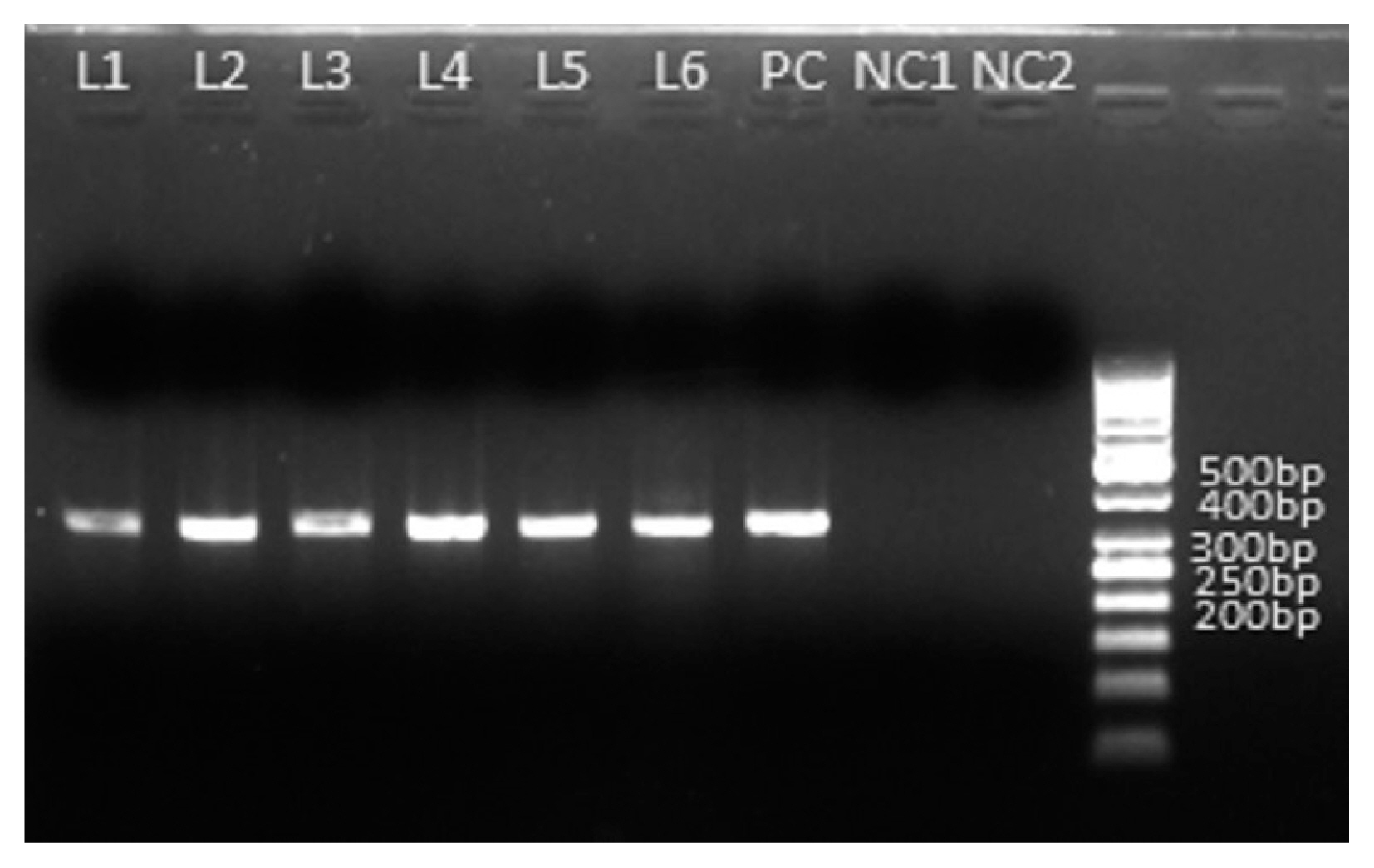
Cryptosporidium spp. isolates identified in and around Kolkata, we employed sequencing of amplified products of HSP70 locus. As the same size amplified products (
Fig. 2) do not unavoidably mean identical DNA sequences, we employed directly the sequencing of the PCR products without cloning them into any vector to reduce the chances of any sequence selection. We successfully sequenced all the positive samples and the obtained sequences were subjected to BLAST analysis after the bidirectional automated sequencing of the PCR product. The BLAST analysis of the sequences confirmed the species of
Cryptosporidium isolates. Sequence analysis revealed the presence of 2 major species
Cryptosporidium hominis and
Cryptosporidium parvum along with them one recently described
Cryptosporidium viatorum has been identified. Overall 2.2% (n/N=6/268; 95% CI 1.0% to 4.8%) and 1.9% (n/N=5/268; 95% CI 0.8% to 4.3%) of samples (
Table 1) were showing 99% to 100% identity with
C. parvum and
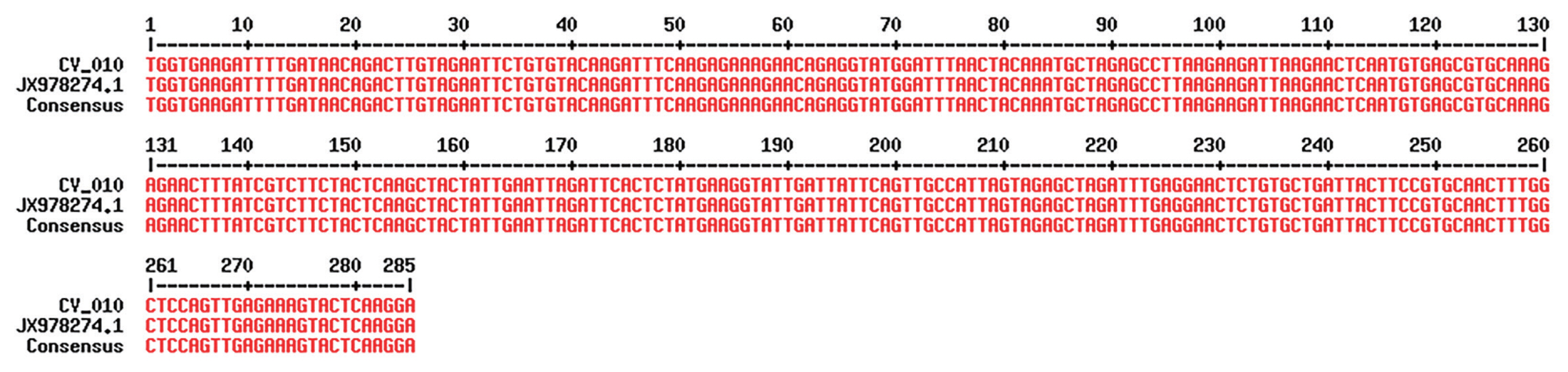
C. hominis. The other species/genotype was detected in only 1 sample (sample ID: PAR/CV_010/2017). Sequencing of the isolate PAR/CV_010/2017 showed complete homology with recently described 3
Cryptosporidium viatorum isolates (GenBank Accession No. JX978274.1, JX978273.1 and JN846706.1) (
Fig. 3).
C. viatorum sequence identified in the present study has been deposited in GenBank under the accession number MT341769 (
Cryptosporidium viatorum HSP70 gene). Overall 0.4% (n/N=1/268; 95% CI 0.0% to 2.1%) of samples (
Table 1) were positive for
C. viatorum and the infected individual was an 11-year-old male patient from Kolkata admitted to I.D. hospital, Kolkata with diarrhoeal complains. The symptoms for
C. viatorum infection were almost similar like diarrhoea, abdominal pain, fever and nausea compared to cases of
C. hominis and
C. parvum except too much vomiting which was reported less often in typical cryptosporidisis. Stool sample from the patient showed the presence of
Cryptosporidium oocytes under microscope after Kinyoun staining, a modified acid-fast staining technique. As with the other
Cryptosporidium species, oocysts of
C. viatorum are indistinguishable from predominantly human-infecting species of
Cryptosporidium. The
C. viatorum isolate was found with no geographical variation in the species that has been described exclusively on the Indian Subcontinent.
C. viatorum is a globally distributed enteric parasite which was identified in many patients travelled to Kenya, Guatemaala and Indian subcontinent.
Cryptosporidium viatorum was first identified in 2012 from travellers returning to Great Britain from Indian subcontinent with gastrointestinal symptoms. However, only 1reports of C. viatorum infection from India are available till date. This report on molecular identification of C. viatorum warrants the necessity of further studies on phylogeography, host specificity, host parasite interaction and clinical importance of the little studied parasite in Indian population as well as in animals.
This report describes 1 unusual
Cryptosporidium species/genotypes involved in human infections in Indian subcontinent. Data from this region are still limited and to the best of our knowledge, this is the second report from India and first report from Bengal on the molecular identification of
C. viatorum infecting humans. Elwin et al. [
5] reported that natural infections with
C. viatorum results diarrhoea, abdominal pain, fever and occasionally nausea that typical for human cryptosporidiosis and that also correspond well with our results.
The morphology and staining characteristics of
C. viatorum oocysts are typical of the genus; particularly for
C. homins and
C. parvum which are the main infective species in human, suggesting that for diagnostic purpose the current microscopical study are suitable but unable to differentiate from other
Cryptosporidium species [
5]. Different types of molecular techniques are being used in differentiation of
Cryptosporidium species/genotypes [
10]. Sequenced data from partial gene loci of HSP70 was applied here for species differentiation.
In India the potential zoonotic transmission of
Cryptosporidium species between cattle and humans has already reported [
11]. Sofar, no natural host has been reported except recently reported non-human host of
C. viatorum in Australian swamp rats and potential reservoir remains unknown; but the possibility of identifying the species in animal hosts must be included as very little is known about
Cryptosporidium spp. in India [
1].
Our data indicates that although the number of reported C. viatorum cases is small, some trends seem to have emerged. As data are being accumulated, risk analysis in cases that potentially involve Cryptosporidium spp. will be improved.
Notes
-
The authors declare that they have no conflicts of interest in publication of this paper.
ACKNOWLEDGMENTS
The study was supported by a grant from National Institute of Infectious Disease, Tokyo, Japan. The authors would like to thank Dr. Koushik Das for his immense help for providing technical support.
Fig. 1Microscopic view of C. viatorum oocysts after modified acid fast staining (×100).

Fig. 2Amplification of HSP70 gene of Cryptosporidium spp. of 6 microscopy positive samples. PCR product size: 324 bpp L1–L6 are amplified PCR products of Cryptosporidium spp. PC is amplified PCR product of Cryptosporidium parvum as a positive control. NC1 and NC2 are negative control contain no stool DNA.

Fig. 3Alignment of 1 local isolates (sample ID: PAR/CV_010/2017) of C. viatorum and reference strain using MultAlign after sequencing; the alignment shows 100% similarity with reference strain.

Table 1Prevalence of different Cryptosporidiums species after PCR assay among the patients admitted to Infectious Disease & Beliaghata General Hospital, Kolkata, India with diarrheal complaints from March 2017 to February 2020
Table 1
|
Species |
PCR |
|
No. examined (N) |
Positive (n) |
Prevalence (% of positive) |
95% CI |
|
Cryptosporidium parvum
|
|
6 |
2.2 |
1.0 to 4.8 |
|
Cryptosporidium hominis
|
268 |
5 |
1.9 |
0.8 to 4.3 |
|
Cryptosporidium viatorum
|
|
1 |
0.4 |
0.0 to 2.0 |
References
REFERENCESS
- 1. Koehler AV, Wang T, Haydon SR, Gasser RB. Cryptosporidium viatorum from the native Australian swamp rat Rattus lutreolus-An emerging zoonotic pathogen? Int J Parasitol Parasites Wildl 2018;7:18-26. https://doi.org/10.1016/j.ijppaw.2018.01.004
- 2. Bamaiyi PH, Redhuan NEM. Prevalence and risk factors for cryptosporidiosis: a global, emerging,neglected zoonosis. Asian Biomedicine 2016;10:309-325. https://doi.org/10.5372/1905-7415.1004.493
- 3. Ayinmode AB, Zhang H, Dada-Adegbola HO, Xiao L. Cryptosporidium hominis subtypes and Enterocytozoon bieneusi genotypes in HIV-infected persons in Ibadan, Nigeria. Zoonoses Public Health 2014;61:297-303. https://doi.org/10.1111/zph.12072
- 4. Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, Feng Y, Xiao L. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl Trop Dis 2014;8:e2831. https://doi.org/10.1371/journal.pntd.0002831
- 5. Elwin K, Hadfield SJ, Robinson G, Crouch ND, Chalmers RM. Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. Int J Parasitol 2012;42:675-682. https://doi.org/10.1016/j.ijpara.2012.04.016
- 6. Lebbad M, Beser J, Insulander M, Karlsson L, Mattsson JG, Svenungsson B, Axén C. Unusual cryptosporidiosis cases in Swedish patients: extended molecular characterization of Cryptosporidium viatorum and Cryptosporidium chipmunk genotype I. Parasitology 2013;140:1735-1740. https://doi.org/10.1017/S003118201300084X
- 7. Peralta RH, Velásquez JN, Cunha Fde S, Pantano ML, Sodré FC, Silva Sd, Astudillo OG, Peralta JM, Carnevale S. Genetic diversity of Cryptosporidium identified in clinical samples from cities in Brazil and Argentina. Mem Inst Oswaldo Cruz 2016;111:30-36. https://doi.org/10.1590/0074-02760150303
- 8. Insulander M, Silverlås C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect 2013;141:1009-1020. https://doi.org/10.1017/S0950268812001665
- 9. Shehla Khalil, Mirdha BR, Paul J, Panda A, Singh Y. Molecular detection and identification of Cyptosporidium viatorum in a human immunodeficiency virus-seropositive patient. J Glob Infect Dis 2018;10:28-29. https://doi.org/10.4103/jgid.jgid_26_17
- 10. Guyot K, Follet-Dumoulin A, Lelièvre E, Sarfati C, Rabodonirina M, Nevez G, Cailliez JC, Camus D, Dei-Cas E. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J Clin Microbiol 2012;39:3472-3480. https://doi.org/10.1128/JCM.39.10.3472-3480.2001
- 11. Rajendran P, Ajjampur SR, Chidambaram D, Kattula D, Rajan DP, Ward H, Kang G. Investigation of potential zoonotic transmission of cryptosporidiosis in southern India. Am J Trop Med Hyg 2011;85:657-659. https://doi.org/10.4269/ajtmh.2011.10-0673
 , Ajanta Ghosal1, Yumiko Saito-Nakano2, Shanta Dutta3, Tomoyoshi Nozaki4, Sandipan Ganguly1,*
, Ajanta Ghosal1, Yumiko Saito-Nakano2, Shanta Dutta3, Tomoyoshi Nozaki4, Sandipan Ganguly1,*