Abstract
Although helminth parasites have different life cycles, their hosts share similar immune responses involving Th2 cell-type. Here, we extracted proteins from the larvae of Anisakis simplex complex and Trichinella spiralis to identify common and specific antigens (or allergens) associated with the Th2 immune response. We performed two-dimensional electrophoresis analysis and Matrix-assisted laser desorption ionization–time of flight/time of flight (MALDI-TOF/TOF) experiments. We found 13 potentially immunogenic proteins, which included 5 spots specific to T. spiralis and 8 common to T. spiralis and A. simplex, by tandem mass spectrometry. These molecules were identified structurally as actin, tropomyosin, col cuticle N domain-containing protein, and heat shock proteins. We also identified molecules related to parasite-host immune modulation and interactions. Our results may contribute to reveal potential roles of immunological proteins in parasite-derived immune modulation.
-
Key words: Proteome, Anisakis simplex, Trichinella spiralis, two-dimensional electrophoresis
Introduction
Helminth parasites have coexisted with their intermediate/definitive hosts for a very long time, during which they have evolved various immune regulatory mechanisms. Helminthic infections typically induce a strong Th2 immune response in the host immune system. In response, the parasite has evolved various immune evasion mechanisms that disrupt the host immune system, thus enabling the parasite to survive attacks from the host immune system. A representative strategy is to activate immune regulatory cells, such as regulatory T (Treg) cells, and regulatory B cells; alternatively, helminth may also activate macrophages that can control the host inflammatory response [
1].
Interestingly, immune cells induced by parasitic infections act as bystanders that suppress the onset and progression of autoimmune diseases or excessive inflammatory responses to allergens. Therefore, the suppression of immune diseases caused by parasitic infections has recently become a hot topic in parasitic research [
2].
Anisakis simplex larvae parasitize the human body and cause anisakiasis. Generally, its life cycle involves stages within the bodies of fish or marine mammals. In humans,
A. simplex infection produces immunoglobulin E (IgE), which can cause allergic reactions such as anaphylaxis [
3]. Meanwhile,
Trichinella spiralis is a nematode that can be transmitted by the consumption of raw or undercooked meat (usually pork) [
4]. Infected patients may experience intestinal problems and myalgia, with an increase in serum IgE and eosinophilia, which are similar to the symptoms of anisakiasis [
4].
We have previously observed a Treg cell-related immune response following an experimental parasite infection [
5]. Although
Anisakis infections mainly elicit allergic reactions in humans, some elicited proteins may have immunomodulatory functions [
6]. Therefore, we hypothesized that similar antigens or allergens may act on the Th2 and Treg cell-related immune responses that different nematode parasites similarly induce in their hosts. By identifying the common and specific proteins of the 2 parasites, we aimed to identify candidate proteins and analyze their functions in host immune regulation during parasite infection. Proteins were identified using MALDI-TOF/TOF.
Materials and Methods
Ethics statement
The mice housed at a specific pathogen-free facility at the Institute for Laboratory Animals of Pusan National University, where all animal studies were conducted. In line with “The Act for the Care and Use of Laboratory Animals” of the Ministry of Food and Drug Safety of Korea, the Pusan National University Animal Care and Use Committee registered and approved the experiments (approval no. PNU-2020-2637).
Materials
Acrylamide (Sigma-Adrich, St. Louis, MO, USA), acetonitrile (Sigma-Adrich), benzamidine (Sigma-Adrich), bis-acrylamide, Bradford,3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate (CHAPS) solution (Sigma-Adrich), dithiothreitol (DTT), iodoacetamide (Sigma-Adrich), sodium dodecyl sulfate (SDS) (Sigma-Adrich), trifluoroacetic acid (Sigma-Adrich), urea (Sigma-Adrich), thiourea (Sigma-Adrich), and α-cyano-4-hydroxy-cinnamic acid (Sigma-Adrich). Pharmalyte (pH 3.5–10) (Amersham, Buckinghamshire, United Kingdom) Immobiline DryStrip gels (IPG DryStrips) (pH 4–10 NL, 24 cm) (Genomine, Korea). Modified porcine trypsin (sequencing-grade) (Promega, Madison, WI, USA).
Parasite preparation
Trichinella spiralis strain ISS623 was maintained in experimental mice by infection. The
T. spiralis muscle larvae were collected as previously reported [
5]. The
A. simplex L3 larvae were collected from infected
Scomber japonicus as previously reported [
7].
Protein sample preparation
The total protein pellets were washed twice with ice-cold Phosphate-buffered saline (PBS) (Sigma-Adrich), and sonicated for 10 s using a Sonoplus (Bandelin electronic, Germany), and then they were homogenized using a motor-driven homogenizer (PowerGen 125, Fisher Scientific) in a sample lysis solution (7 M urea, 2 M thiourea, 1% (w/v) dithiothreitol (DTT), 2% (v/v) pharmalyte, 4% (w/v) 3-[(3-cholamidopropy) dimethyammonio]-1-propanesulfonate (CHAPS), and 1 mm benzamidine). Homogenized samples were subjected to freezing and thawing steps 5 times in one day. Occasionally, a bead beater was used to lyse rigid cells. Proteins from pellets were extracted by vortex mixing for 1 h at room temperature. Samples were then centrifuged at 15,000×g for one hour at 15ºC, and the insoluble material was discarded. The soluble fraction was analyzed by 2-dimensional gel electrophoresis as described below. Protein concentration was determined using the Bradford method [
5].
Immobilized pH gradient (IPG) dry strips were equilibrated for 12–16 h with equal volumes of 7 M urea and 2 M thiourea containing 2% CHAPS, 1% DTT, and 1% pharmalyte. Six hundred μg of each sample was then loaded on the gel. Isoelectric focusing (IEF) was performed at 20ºC using a Multiphor II electrophoresis unit with an EPS 3,500 XL power supply (Amersham Biosciences) following the manufacturer’s instructions. IEF conditions were as follows: the voltage was linearly increased from 150 V to 3,500 V over 3 h, followed by a constant 3,500 V, with complete focusing after 96 kVh. Prior to the second-dimension run, the strips were incubated for 10 min each in equilibration buffers (50 mm Tris-Cl, pH 6.8 containing 6 M urea, 30% glycerol and 2% SDS) containing 1% DTT and then 2.5% iodoacetamide. Equilibrated strips were inserted into SDS-PAGE gels (20 cm×24 cm, 10–16%). SDS-PAGE was performed using a Hoefer DALT 2D system (Amersham Biosciences) following the manufacturer’s instruction. The 2D gels were run at 20ºC for 1,700 Vh, followed by staining with Coomassie G250.
In-gel protein digestion
The protein spots were enzymatically digested in-gel using porcine trypsin. The gel pieces were first washed with 50% acetonitrile to remove SDS, salt, and stain. The washed and dehydrated spots were then vacuum dried to remove solvent and rehydrated with trypsin (8–10 ng/μl) solution in 50 mm ammonium bicarbonate pH 8.7 and incubated for 8–10 h at 37ºC.
Identification of proteins by MALDI-TOF/TOF
The samples were analyzed using a BRUKER Autoflex maX instrument with LIFT ion optics. Both MS and MS/MS data were acquired using a SMARTBEAM LASER with a 2 kHz repetition rate, and up to 4,000 shots were accumulated for each spectrum. The MS/MS mode was operated with 2 keV collision energy, and air was used as the collision gas to achieve nominal single collision conditions. We used a resolution of 100 in this study. Both MS and MS/MS data were acquired using the instrument’s default calibration without applying internal or external calibration. The MS/MS ion searches were performed using our in-house Mascot license.
Results
2-DE reference map profile of T. spiralis and A. simplex proteins
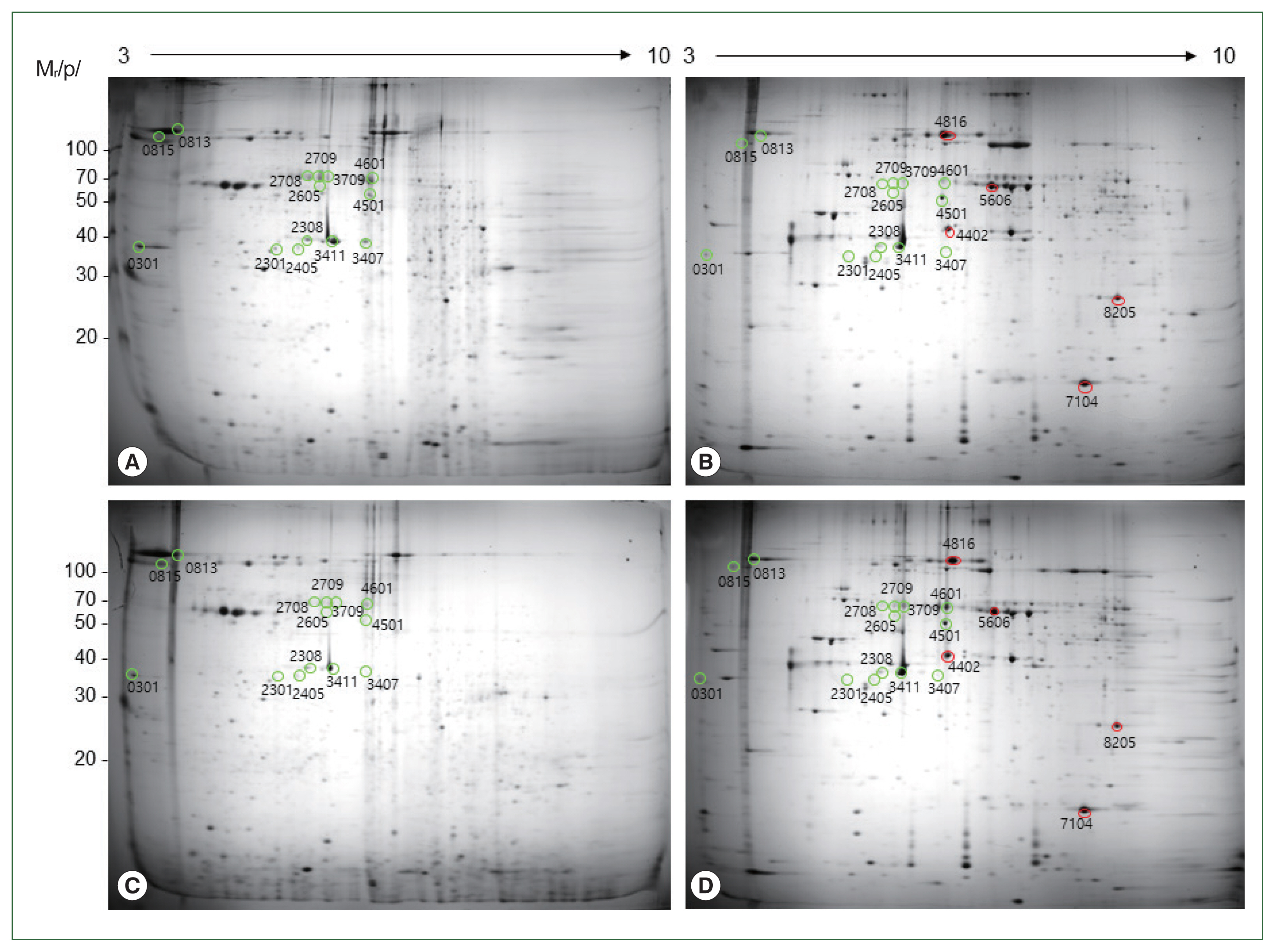
The 2-DE lysis solution was directly added to the sample to quantify the extracted protein. Six hundred microgram of sample were applied to the gel then stained with Colloidal Coomassie Brilliant Blue. Detailed images are listed below by enlarging what is considered to be a spot that exists in a similar location and a similar shape. We selected 14 spots with similar locations and shapes. We also selected 5 spots that were specific to
T. spiralis (
Fig. 1). In addition, we selected 19 abundant spots based on their normalized volumes, and then we processed these spots for further identification by peptide mass fingerprinting (PMF) using MALDI-TOF MS.
The putative protein spots were excised from the gels, digested in-gel by trypsin, and analyzed by MALDI-TOF MS. From a total of 19 spots, we were able to identify 13 proteins, including 5 that were specific to
T. spiralis (
Table 1) and 8 that were common to x (
Tables 2,
3). Three of the 14 common spots (spots 2,708, 2,709, and 3,709) were identified as
H. sapiens proteins, and are likely the result of contamination during protein isolation. We were unable to identify 3 (spots 813, 4,601, and 2,605) of the 14 common spots. One spot (4,816) of
T. spiralis was identified as a protein in
Hydra vulgaris. Five
T. spiralis-specific proteins were identified as: heat shock protein 70, a serine protease, 32-kDa beta-galactoside-binding lectin (Galectin-1), small heat shock protein, and uncharacterized protein LOC100211249. Common spots 3,411 and 2,308 in
A. simplex and
T. spiralis were identified as actin. Spots 2,405 and 3,407 were not identified in
T. spiralis but were identified as actin in
A. simplex. Spot 4,501 was identified as heat shock protein (60 kDa). Common spots 301 in
A. simplex and
T. spiralis were identified as tropomyosin and calumenin A, respectively. Common spots 815 and 2,405 were not identified in
T. spiralis, but they were identified as col cuticle N-domain-containing proteins and actin, respectively, in
A. simplex.
Discussion
Helminth proteins have the capacity to modulate the immune response.
T. spiralis induced a type 2 immune response in a model of inflammatory bowel disease and diet induced obesity with an excessive increase in type 1 immune response, attenuating its symptoms. In addition, nematode infection or treatment with
T. spiralis-derived substances suppresses allergic respiratory inflammatory responses. Our previous studies reported that
T. spiralis can improve the effects of obesity [
8,
9]. Moreover,
A. simplex recombinant protein alleviates allergic airway inflammation [
6]. Here, our aim was to identify, from the larvae of 2 parasites, immunomodulatory substances that alleviate immune disease symptoms.
The main problem for proteomic analysis of parasitic nematodes is the lack of available genomic data that can be used to analyze mass spectrometry data. Optimization of the 2-DE proteomic profiles of A. simplex and T. spiralis is necessary for the benefit of future investigations of proteomic biomarkers and novel candidate proteins involved in host immune regulation. Therefore, we generated 2-DE reference maps of proteins, and our resulting gel images were processed to select 19 highly expressed proteins for MALDI-TOF MS analysis.
We identified 5 spots that are specific to T. spiralis as serine protease, Galectin-1, small heat shock protein, heat shock protein 70, and uncharacterized protein LOC100211249. However, only the first 4 proteins are associated with T. spiralis, while the uncharacterized protein LOC100211249 belongs to Hydra vulgaris.
Serine proteases are recognized by the host immune system during natural infection as immunodominant antigens [
10]. Additionally, inhibiting serine protease activity can prevent inflammation and insulin resistance induced by a high-fat diet [
11]. Serine protease is a crucial enzyme involved in both the onset and progression of inflammation. Because an altered immune response to nematode infection can attenuate inflammation in inflammatory bowel diseases and allergic airway inflammation, we speculated that certain
T. spiralis-specific proteins may be able to suppress inflammation. Research on the association between serine protease and inflammatory bowel diseases, such as Crohn’s disease and ulcerative colitis, is extensive [
12]. A previous report suggests that inhibiting serine protease activity can suppress inflammation, which may be a promising approach for treatment and prevention strategies for inflammatory bowel diseases [
13]. Moreover, serine protease plays a key role in respiratory inflammation, i.e., it regulates cytokines and chemicals that promote inflammation and cause mucosal tissue damage in the airways [
14].
Galectin-1 of
T. spiralis adult worm has been detected in the sera of infected pigs and mice at 7 day post infection (dpi) [
10]. Galectin-1 is a protein capable of binding carbohydrates and is expressed in several cell types in the body. It plays key roles in tissue maturation, remodeling, and homeostasis through the regulation of cell angiogenesis, migration, proliferation, apoptosis, and inflammation [
15]. Galectin-1 gene expression is elevated in the adipocytes of type 2 diabetes patients and obese mice [
16]. Additionally, several research groups have reported an association between Galectin-1 expression and the metabolic outcomes of adipose tissue in various animal models [
17]. Galectin-1 induces the suppressive phenotype in immune cells, recruits immunosuppressive cells, and impairs the function of cytotoxic leukocytes. Recent studies have revealed that Galectin-1 plays a proinflammatory role in certain diseases [
18], as well as in allergic respiratory diseases such as allergic rhinitis, asthma, and chronic obstructive pulmonary disease (COPD) [
19]. It regulates the immune and inflammatory responses by impacting cell interactions, balancing inflammatory cell populations, and regulating cell death [
20]. Galectin-1 plays an important role in regulating inflammation and immune responses associated with inflammatory bowel diseases. This protein is also involved in modulating inflammatory responses and controlling tissue damage. Specifically, Galectin-1 plays roles in regulating the expression of proinflammatory cytokines and inhibiting inflammation [
21].
LOC100211249 is an uncharacterized protein of Hydra vulgaris. A Hydra is a freshwater polyp belonging to the Cnidaria. Its anatomy consists of a tube with 2 cell layers and 3 stem cell populations along its oral-peritoneal axis. H. vulgaris is a small freshwater hydroid with a length ranging from 10 to 30 mm and width of about 1 mm.
Reciprocal Best Hits (RBH) is a common proxy for orthology in comparative genomics. RBH analyses were performed. Proteomes of
T. spiralis were obtained from UniProtKB (
https://www.uniprot.org/help/uniprotkb) [
22]. We identified 2
T. spiralis-specific proteins as small heat shock protein and HSP70. HSP60 was found in both parasites. Among the proteins common to both
A. simplex and
T. spiralis, the most common one was actin, which is one of the most abundant proteins in eukaryotic cells. Actin can polymerize into filaments (F-actin) and form static or highly dynamic networks that play roles in many crucial cellular processes [
23]. In the case of respiratory allergies, actin may play a role in the inflammatory response of the respiratory organs reacting to external stimuli and potentially triggering symptoms of allergies or other respiratory conditions [
24]. In inflammatory bowel diseases, such as colitis, Crohn’s disease, and ulcerative colitis, actin contributes to maintaining the structure of intestinal cells and regulating intestinal movements. The inflammatory processes within the intestines in these conditions may be due to abnormalities in cell movement related to actin [
25]. Yang et al. [
26] showed that the immunoproteomic profile of
T. spiralis adult worms consists of actin and heat shock protein, and that these proteins play roles in the invasion of the worm into intestinal epithelial cells, indicating that they protein may be involved in the early survival of parasites within the host. Filarial tropomyosin shares structural features and cross-reacts with B-cell epitopes of other highly allergenic invertebrate tropomyosins. Recent data have described mechanisms that may prevent hosts from developing allergic responses against allergens of their parasites, such as nematodes or filarial tropomyosins [
27]. Several tropomyosin vaccine studies have been conducted using various nematodes [
28]. Actin plays an important role in linking the complex signaling pathways and cytoskeletal elements of T-cells undergoing activation. Actin and actin-related proteins also regulate T-cell activation [
29]. Tropomyosin is a microfilament-related protein present in all eukaryotic cells [
27]. Heat shock proteins (HSPs) are a highly conserved and immunogenic family of proteins that may function as immunological modulators, and they are potential vaccine antigen candidates [
30]. HSPs are implicated in several immunomodulatory functions. Interestingly, studies on the effects of some HSPs on immune cell function have produced contradictory results, which appear to depend on the protein concentration. For instance, reduced levels of HSP60 prompt anti-inflammatory responses, while elevated levels trigger proinflammatory responses [
31]. Additionally, HSP70 serves as a primary target for host immune responses against infections caused by helminths and protozoan parasites [
32]. Earlier research has suggested that SmHSP70 triggers an early humoral immune response and may be a promising target for the immunodiagnosis of schistosomiasis [
33]. HSP70 in
Echinostoma caproni induces a strong early immune response in mice [
34]. Furthermore, Ts-Hsp70 from adult
T. spiralis is a candidate vaccine because of its high immunogenicity [
35]. Heat shock proteins may play a role in promoting or dampening allergic inflammation, e.g., certain HSPs induce the production of proinflammatory cytokines that tend to exacerbate allergic responses. In contrast, other HSPs are anti-inflammatory and can attenuate allergic inflammation [
36]. The col cuticle N-domain is located in the N-terminal region of the nematode cuticle collagen, and its function remains unknown. The cuticle is a tough elastic structure secreted by hypodermal cells and primarily consists of collagen proteins [
37].
Contrary to our expectations of identifying proteins related to immune regulation, our analysis of common spots in 2 parasites yielded unexpected results. Our findings indicate that the role of shared molecules among parasites in immunity remains unresolved and warrants further investigation.
Notes
-
Author contributions
Conceptualization: Kang SA, Yu HS
Data curation: Kang SA
Formal analysis: Kang SA
Funding acquisition: Yu HS
Investigation: Kang SA
Methodology: Kang SA
Writing – original draft: Kang SA
Writing – review & editing: Yu HS
-
The authors declare no conflict of interest related to this study.
Acknowledgment
This work was supported by a 2-Year Research Grant of Pusan National University.
Fig. 1Colloidal Coomassie blue-stained 2-DE gel images of T. spiralis and A. simplex proteomes. The T. spiralis and A. simplex proteins (300 μg each) were fractionated on pH 3–10 non-linear immobilized pH gradient strips, after which run on a 12.5% polyacrylamide gel. A total of 100 spots with the highest normalized volumes (area×intensity) were analyzed using matrix assisted laser desorption ionization-time of flight mass (MALDI-TOF MS) with Peptide mass fingerprinting (PMF) searching of databases. Nineteen spots were analyzed using Q-TOF MS/MS with Ion Searches of the putative EST database. Sequences identified in this manner were used for BLASTp searches of the NCBI protein database. (A, B) Anisakis simplex, (C, D) Trichinella spiralis. Green spots are those common to both parasites (Spots 3417, 4501, 2308, 301, 2301, 815, 2405, 3407, 2708, 2709, 3709, 813, 4601, and 2605); red circle indicates spots specific to T. spiralis (Spots 4402, 8205, 7104, 5606, and 4,816).

Table 1Protein identification of T. spiralis specific 5 spots with the highest Mascot scores picked on deep purple stained 2D-gel
Table 1
|
Spot no. |
Accession no. |
Description |
Nominal mass (Mr) |
Score |
Sequence coverage (%) |
Calculated pI |
Taxonomy |
|
4,402 |
Gi|168805931 |
Serine protease |
35,704 |
256 |
15 |
5.97 |
Trichinella spiralis
|
|
8,205 |
Gi|316970057 |
32 kDa beta-galactoside-binding lectin (Galectin-1) |
55,069 |
322 |
17 |
9.04 |
Trichinella spiralis
|
|
7,104 |
Gi|116090563 |
Small heat shock protein |
18,935 |
127 |
27 |
6.32 |
Trichinella spiralis
|
|
5,606 |
Gi|152004108 |
Heat shock protein 70 |
71,243 |
282 |
12 |
5.48 |
Trichinella nativa
|
|
4,816 |
Gi|449665026 |
PREDICTED: uncharacterized protein LOC100211249 |
93,289 |
16 |
2 |
8.90 |
Hydra vulgaris
|
Table 2Protein identification of A. simplex of common spots with the highest Mascot scores picked on deep purple stained 2D-gel
Table 2
|
Spot no. |
Accession no. |
Description |
Nominal mass (Mr) |
Score |
Sequence coverage (%) |
Calculated pI |
Taxonomy |
|
3,411 |
A0A0B4SV59 |
Actin |
42,132 |
572 |
28 |
5.30 |
Anisakis simplex
|
|
4,501 |
A0A0M3K0Q9 |
Chaperonin homolog Hsp-60, mitochondrial |
50,811 |
203 |
8 |
4.99 |
Anisakis simplex
|
|
2,308 |
A0A0B4SV59 |
Actin |
42,132 |
601 |
30 |
5.30 |
Anisakis simplex
|
|
301 |
A0A0M3KCE6 |
Tropomyosin |
13,693 |
57 |
13 |
4.54 |
Anisakis simplex
|
|
2,301 |
A0A0B4SV59 |
Actin |
42,132 |
47 |
2 |
5.30 |
Anisakis simplex
|
|
815 |
A0A0M3JVZ5 |
Col cuticle N domain-containing protein |
28,549 |
81 |
10 |
8.51 |
Anisakis simplex
|
|
2,405 |
A0A0B4SV59 |
Actin |
42,132 |
70 |
4 |
5.30 |
Anisakis simplex
|
|
3,407 |
A0A0B4SV59 |
Actin |
42,132 |
168 |
10 |
5.30 |
Anisakis simplex
|
Table 3Protein identification of T. spiralis of common spots with the highest Mascot scores picked on deep purple stained 2D-gel
Table 3
|
Spot no. |
Accession no. |
Description |
Nominal mass (Mr) |
Score |
Sequence coverage (%) |
Calculated pI |
Taxonomy |
|
3,411 |
E5SLU1 |
Actin-5C |
42,210 |
340 |
15 |
5.30 |
Trichinella spiralis
|
|
4,501 |
A0A0V1BDW1 |
60 kDa heat shock protein, mitochondrial |
158,326 |
338 |
5 |
6.55 |
Trichinella spiralis
|
|
2,308 |
A0A0V1C146 |
Actin-5C |
41,189 |
223 |
12 |
5.22 |
Trichinella spiralis
|
|
301 |
A0A0V1B5E1 |
Calumenin-A |
36,465 |
104 |
6 |
4.51 |
Trichinella spiralis
|
|
2,301 |
A0A0V1BML4 |
Malate dehydrogenase (Fragment) |
108,271 |
30 |
1 |
6.14 |
Trichinella spiralis
|
References
- 1. Maizels RM, McSorley HJ. Regulation of the host immune system by helminth parasites. J Allergy Clin Immunol 2016;138(3):666-675.
https://doi.org/10.1016/j.jaci.2016.07.007
- 2. McSorley HJ, Maizels RM. Helminth infections and host immune regulation. Clin Microbiol Rev 2012;25(4):585-608.
https://doi.org/10.1128/CMR.05040-11
- 3. Polak I, Stryiński R, Majewska M, Łopieńska-Biernat E. Metabolomic analysis reveals a differential adaptation process of the larval stages of Anisakis simplex to the host environment. Front Mol Biosci 2023;10:1233586.
https://doi.org/10.3389/fmolb.2023.1233586
- 4. Diaz JH, Warren RJ, Oster MJ. The disease ecology, epidemiology, clinical manifestations, and management of trichinellosis linked to consumption of wild animal meat. Wilderness Environ Med 2020;31(2):235-244.
https://doi.org/10.1016/j.wem.2019.12.003
- 5. Kang SA, Cho MK, Park MK, Kim DH, Hong YC, et al. Alteration of helper T-cell related cytokine production in splenocytes during Trichinella spiralis infection. Vet Parasitol 2012;186(3–4):319-327.
https://doi.org/10.1016/j.vetpar.2011.12.002
- 6. Cho MK, Park MK, Kang SA, Park SK, Lyu JH, et al. TLR2-dependent amelioration of allergic airway inflammation by parasitic nematode type II MIF in mice. Parasite Immunol 2015;37(4):180-191.
https://doi.org/10.1111/pim.12172
- 7. Kim J, Jo JO, Choi SH, Cho MK, Yu HS, et al. Seroprevalence of antibodies against Anisakis simplex larvae among health-examined residents in three hospitals of southern parts of Korea. Korean J Parasitol 2011;49(2):139-144.
https://doi.org/10.3347/kjp.2011.49.2.139
- 8. Kang SA, Yu HS. Anti-obesity effects by parasitic nematode (Trichinella spiralis) total lysates. Front Cell Infect Microbiol 2023;13:1285584.
https://doi.org/10.3389/fcimb.2023.1285584
- 9. Kang SA, Choi JH, Baek KW, Lee DI, Jeong MJ, et al.
Trichinella spiralis infection ameliorated diet-induced obesity model in mice. Int J Parasitol 2021;51(1):63-71.
https://doi.org/10.1016/j.ijpara.2020.07.012
- 10. Yang J, Pan W, Sun X, Zhao X, Yuan G, et al. Immunoproteomic profile of Trichinella spiralis adult worm proteins recognized by early infection sera. Parasit Vectors 2015;8:20.
https://doi.org/10.1186/s13071-015-0641-8
- 11. Kuo CS, Chen JS, Lin LY, Schmid-Schonbein GW, Chien S, et al. Inhibition of Serine protease activity protects against high fat diet-induced inflammation and insulin resistance. Sci Rep 2020;10(1):1725.
https://doi.org/10.1038/s41598-020-58361-4
- 12. Kriaa A, Jablaoui A, Mkaouar H, Akermi N, Maguin E, et al. Serine proteases at the cutting edge of IBD: focus on gastrointestinal inflammation. Faseb J 2020;34(6):7270-7282.
https://doi.org/10.1096/fj.202000031RR
- 13. Patel S. A critical review on serine protease: key immune manipulator and pathology mediator. Allergol Immunopathol (Madr) 2017;45(6):579-591.
https://doi.org/10.1016/j.aller.2016.10.011
- 14. Soh WT, Zhang J, Hollenberg MD, Vliagoftis H, Rothenberg ME, et al. Protease allergens as initiators-regulators of allergic inflammation. Allergy 2023;78(5):1148-1168.
https://doi.org/10.1111/all.15678
- 15. Wang J, Zhang S, Wang Y, Zhu Y, Xu X, et al. Effect of galectin-1 on prognosis and responsiveness of immune checkpoint plus tyrosine kinase inhibition in renal cell carcinoma. Cancer Med 2024;13(7):e7113.
https://doi.org/10.1002/cam4.7113
- 16. Baek JH, Kim DH, Lee J, Kim SJ, Chun KH. Galectin-1 accelerates high-fat diet-induced obesity by activation of peroxisome proliferator-activated receptor gamma (PPARγ) in mice. Cell Death Dis 2021;12(1):66.
https://doi.org/10.1038/s41419-020-03367-z
- 17. Fryk E, Silva VRR, Jansson PA. Galectin-1 in obesity and type 2 diabetes. Metabolites; 2022. 12(10):
https://doi.org/10.3390/metabo12100930
- 18. Yu X, Qian J, Ding L, Yin S, Zhou L, et al. Galectin-1: a traditionally immunosuppressive protein displays context-dependent capacities. Int J Mol Sci; 2023. 24(7):
https://doi.org/10.3390/ijms24076501
- 19. Lv Y, Dai M, Wang M, Chen F, Liu R. Anti-inflammatory property of galectin-1 in a murine model of allergic airway inflammation. J Immunol Res 2019;2019:9705327.
https://doi.org/10.1155/2019/9705327
- 20. Ge XN, Ha SG, Greenberg YG, Rao A, Bastan I, et al. Regulation of eosinophilia and allergic airway inflammation by the glycan-binding protein galectin-1. Proc Natl Acad Sci U S A 2016;113(33):4837-4846.
https://doi.org/10.1073/pnas.1601958113
- 21. Arda-Pirincci P, Aykol-Celik G. Galectin-1 reduces the severity of dextran sulfate sodium (DSS)-induced ulcerative colitis by suppressing inflammatory and oxidative stress response. Bosn J Basic Med Sci 2020;20(3):319-328.
https://doi.org/10.17305/bjbms.2019.4539
- 22. Wenger Y, Galliot B. RNAseq versus genome-predicted transcriptomes: a large population of novel transcripts identified in an Illumina-454 Hydra transcriptome. BMC Genomics 2013;14:204.
https://doi.org/10.1186/1471-2164-14-204
- 23. Das S, Stortz JF, Meissner M, Periz J. The multiple functions of actin in apicomplexan parasites. Cell Microbiol 2021;23(11):e13345.
https://doi.org/10.1111/cmi.13345
- 24. Mikami M, Yocum GT, Heller NM, Emala CW. Reduced allergic lung inflammation and airway responsiveness in mice lacking the cytoskeletal protein gelsolin. Am J Physiol Lung Cell Mol Physiol 2020;319(5):833-842.
https://doi.org/10.1152/ajplung.00065.2020
- 25. Lechuga S, Ivanov AI. Actin cytoskeleton dynamics during mucosal inflammation: a view from broken epithelial barriers. Curr Opin Physiol 2021;19:10-16.
https://doi.org/10.1016/j.cophys.2020.06.012
- 26. Wang ZQ, Wang L, Cui J. Proteomic analysis of Trichinella spiralis proteins in intestinal epithelial cells after culture with their larvae by shotgun LC-MS/MS approach. J Proteomics 2012;75(8):2375-2383.
https://doi.org/10.1016/j.jprot.2012.02.005
- 27. Sereda MJ, Hartmann S, Lucius R. Helminths and allergy: the example of tropomyosin. Trends Parasitol 2008;24(6):272-278.
https://doi.org/10.1016/j.pt.2008.03.006
- 28. Hewitson JP, Maizels RM. Vaccination against helminth parasite infections. Expert Rev Vaccines 2014;13(4):473-487.
https://doi.org/10.1586/14760584.2014.893195
- 29. Sun J, Zhong X, Fu X, Miller H, Lee P, et al. The actin regulators involved in the function and related diseases of lymphocytes. Front Immunol 2022;13:799309.
https://doi.org/10.3389/fimmu.2022.799309
- 30. Colaco CA, Bailey CR, Walker KB, Keeble J. Heat shock proteins: stimulators of innate and acquired immunity. Biomed Res Int 2013;2013:461230.
https://doi.org/10.1155/2013/461230
- 31. Zininga T, Ramatsui L, Shonhai A. Heat shock proteins as immunomodulants. Molecules 2018;23(11):https://doi.org/10.3390/molecules23112846.
- 32. Kaur J, Kaur S. ELISA and western blotting for the detection of Hsp70 and Hsp83 antigens of Leishmania donovani.
. J Parasit Dis 2013;37(1):68-73.
https://doi.org/10.1007/s12639-012-0133-0
- 33. Kanamura HY, Hancock K, Rodrigues V, Damian RT. Schistosoma mansoni heat shock protein 70 elicits an early humoral immune response in S. mansoni infected baboons. Mem Inst Oswaldo Cruz 2002;97(5):711-716.
https://doi.org/10.1590/s0074-02762002000500022
- 34. Sotillo J, Valero L, Sanchez Del Pino MM, Fried B, Esteban JG, et al. Identification of antigenic proteins from Echinostoma caproni (Trematoda) recognized by mouse immunoglobulins M, A and G using an immunoproteomic approach. Parasite Immunol 2008;30(5):271-279.
https://doi.org/10.1111/j.1365-3024.2007.01019.x
- 35. Fang L, Sun L, Yang J, Gu Y, Zhan B, et al. Heat shock protein 70 from Trichinella spiralis induces protective immunity in BALB/c mice by activating dendritic cells. Vaccine 2014;32(35):4412-4419.
https://doi.org/10.1016/j.vaccine.2014.06.055
- 36. Shevchenko M, Servuli E, Albakova Z, Kanevskiy L, Sapozhnikov A. The role of heat shock protein 70 kDa in Asthma. J Asthma Allergy 2020;13:757-772.
https://doi.org/10.2147/jaa.S288886
- 37. Van der Eycken W, de Almeida Engler J, Van Montagu M, Gheysen G. Identification and analysis of a cuticular collagen-encoding gene from the plant-parasitic nematode Meloidogyne incognita.
. Gene 1994;151(1–2):237-242.
https://doi.org/10.1016/0378-1119(94)90663-7