1College of Veterinary Medicine and Research Institute for Veterinary Science, Seoul National University, Seoul 08826, Korea
2Department of Laboratory Animal Medicine, College of Veterinary Medicine, Konkuk University, Seoul 05029, Korea
© 2025 The Korean Society for Parasitology and Tropical Medicine
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (https://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Author contributions
Conceptualization: Cho K, Choi YK, Yoo WG
Data curation: Cho K
Funding acquisition: Choi YK, Yoo WG
Investigation: Cho K
Methodology: Cho K, Kim M
Project administration: Choi YK, Yoo WG
Software: Cho K
Supervision: Yoo WG
Validation: Kim M
Visualization: Cho K
Writing – original draft: Cho K, Yoo WG
Writing – review & editing: Kim M, Choi YK, Yoo WG
Conflict of interest
The authors declare no conflict of interest related to this study.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (Ministry of Science and ICT) (grant No. RS-2022-NR070067 and RS-2024-00509361) (http://www.nrf.re.kr) and was partially supported by the Research Institute for Veterinary Science, Seoul National University.
| Gene/region | Positions and size (bp) | Initiation and termination codons | Anticodon | ||||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| Hd-c1a | Hd-p | Hd-c2 | Hd-c1 | Hd-p | Hd-c2 | Hd-c1 | |
| cox3 | 1–651 (651) | 130–780 (651) | 7,122–7,772 (651) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
|
|||||||
| tRNA-His (H) | 660–731 (72) | 789–860 (72) | 7,781–7,852 (72) | GTG | |||
|
|
|||||||
| cytb | 735–1,832 (1,098) | 864–1,961 (1,098) | 7,856–8,953 (1,098) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
|
|||||||
| nad4L | 1,836–2,096 (261) | 1,965–2,225 (261) | 8,957–9,217 (261) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
|
|||||||
| nad4 | 2,063–3,310 (1,248) | 2,192–3,439 (1,248) | 9,202–10,431 (1,230) | ATG/TAG | ATG/TAG | ATT/TAG | |
|
|
|||||||
| tRNA-Gln (Q) | 3,312–3,377 (66) | 3,441–3,505 (65) | 10,433–10,498 (66) | TTG | |||
|
|
|||||||
| tRNA-Phe (F) | 3,377–3,439 (63) | 3,506–3,568 (62) | 10,498–10,560 (63) | GAA | |||
|
|
|||||||
| tRNA-Met (M) | 3,436–3,499 (64) | 3,565–3,628 (64) | 10,577–10,620 (64) | CAT | |||
|
|
|||||||
| atp6 | 3,468–4,019 (552) | 3,633–4,148 (516) | 10,625–11,140 (516) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
|
|||||||
| nad2 | 4,026–4,907 (882) | 4,155–5,036 (882) | 11,147–12,028 (882) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
|
|||||||
| tRNA-Val (V) | 4,908–4,973 (66) | 5,037–5,102 (66) | 12,029–12,093 (65) | TAC | |||
|
|
|||||||
| tRNA-Ala (A) | 4,974–5,043 (70) | 5,103–5,172 (70) | 12,095–12,164 (70) | TGC | |||
|
|
|||||||
| tRNA-Asp (D) | 5,048–5,109 (62) | 5,177–5,238 (62) | 12,169–12,230 (62) | GTC | |||
|
|
|||||||
| nad1 | 5,110–6,000 (891) | 5,239–6,129 (891) | 12,231–13,121 (891) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
|
|||||||
| tRNA-Asn (N) | 6,009–6,072 (64) | 6,138–6,201 (64) | 13,130–13,193 (64) | GTT | |||
|
|
|||||||
| tRNA-Pro (P) | 6,081–6,142 (62) | 6,210–6,271 (62) | 13,202–13,264 (63) | TGG | |||
|
|
|||||||
| tRNA-Ile (I) | 6,142–6,205 (64) | 6,271–6,334 (64) | 13,264–13,325 (62) | GAT | |||
|
|
|||||||
| tRNA-Lys (K) | 6,206–6,269 (64) | 6,335–6,398 (64) | 13,327–13,390 (64) | CTT | |||
|
|
|||||||
| nad3 | 6,273–6,620 (348) | 6,402–6,749 (348) | 13,394–13,741 (348) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
|
|||||||
| tRNA-Ser (S1) | 6,626–6,684 (59) | 6,755–6,813 (59) | 13,747–13,805 (59) | GCT | |||
|
|
|||||||
| tRNA-Trp (W) | 6,685–6,750 (66) | 6,814–6,879 (66) | 13,806–13871 (66) | TCA | |||
|
|
|||||||
| cox1 | 6,754–8,346 (1,593) | 6,883–8,475 (1,593) | 4–1,555 (1,552) | ATG/TAG | ATG/TAG | TTT/ATTb | |
|
|
|||||||
| tRNA-Thr (T) | 8,337–8,401 (65) | 8,466–8,530 (65) | 1,557–1,621 (65) | TGT | |||
|
|
|||||||
| rrnL | 8,402–9,368 (967) | 8,531–9,497 (967) | 1,622–2,588 (967) | ||||
|
|
|||||||
| tRNA-Cys (C) | 9,369–9,435 (67) | 9,498–9,564 (67) | 2,589–2,655 (67) | GCA | |||
|
|
|||||||
| rrnS | 9498–10,137 (640) | 9,565–10,279 (715) | 2,656–3,364 (709) | ||||
|
|
|||||||
| cox2 | 10,151–10,729 (579) | 10,280–10,858 (579) | 3,371–3,949 (579) | ATG/TAA | ATG/TAA | ATG/TAA | |
|
|
|||||||
| tRNA-Glu (E) | 10,730–10,794 (65) | 10,859–10,923 (65) | 3,950–4,014 (65) | ||||
|
|
|||||||
| nad6 | 10,798–11,256 (459) | 10,927–11,385 (459) | 4,018–4,476 (459) | ATG/TAA | ATG/TAA | ATG/TAA | GTA |
|
|
|||||||
| tRNA-Tyr (Y) | 11,260–11,325 (66) | 11,389–11,454 (66) | 4,480–4,545 (66) | TAG | |||
|
|
|||||||
| tRNA-Ser (S2) | 11,511–11,573 (63) | 11,640–11,702 (63) | 4,729–4,795 (67) | NGA | |||
|
|
|||||||
| tRNA-Leu (L1) | 11,588–11,655 (68) | 11,717–11,784 (68) | 4,808–4,875 (68) | TAA | |||
|
|
|||||||
| tRNA-Leu (L2) | 11,681–11,743 (63) | 11,810–11,872 (63) | 4,902–4,964 (63) | TCG | |||
|
|
|||||||
| tRNA-Arg (R) | 11,754–11,813 (60) | 11,883–11,942 (60) | 4,975–5,034 (60) | ||||
|
|
|||||||
| nad5 | 11,808–13,391 (1,584) | 11,946–13,520 (1,575) | 5,038–6,612 (1,575) | ATG/TAG | ATG/TAG | ATG/TAG | TCC |
|
|
|||||||
| tRNA-Gly (G) | 14,025–14,087 (63) | 74–134 (61) | 7,056–7,118 (63) | TTC | |||
|
|
|||||||
| Non-coding region | 13,088–14,090 (643) | 13,521–73 (261) | 6,612–7,055 (444) | ||||
|
|
|||||||
| Repeat unit | 13,495–14,007 (513) | na | 6,612–6,983 (372) | ||||
| Species/strainsa | mPCGs | MRGs | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||||
| atp6 | cox1 | cox2 | cox3 | cytb | nad1 | nad2 | nad3 | nad4L | nad4 | nad5 | nad6 | con-mPCGs | rrnL | rrnS | con-MRGs | |
| H. diminuta (AP017664, Hd-p) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
|
||||||||||||||||
| H. diminuta (NC_002767, Hd-c2) | 0.39 | 0.19 | 0.35 | 0.15 | 0.09 | 0.11 | 0.11 | 0.00 | 0.77 | 0.24 | 0.25 | 0.66 | 0.23 | 0.10 | 0.16 | 0.12 |
|
|
||||||||||||||||
| Hymenolepis nana (NC_029245) | 33.07 | 23.99 | 32.61 | 38.50 | 26.62 | 25.61 | 32.95 | 30.84 | 24.77 | 37.81 | 39.82 | 42.16 | 32.05 | 25.59 | 16.44 | 21.80 |
|
|
||||||||||||||||
| Taenia saginata (NC_009938) | 39.03 | 26.79 | 34.89 | 40.32 | 32.51 | 30.62 | 39.73 | 46.18 | 36.46 | 42.18 | 45.34 | 53.04 | 37.38 | 34.88 | 28.03 | 31.99 |
|
|
||||||||||||||||
| Taenia asiatica (NC_004826) | 40.02 | 26.60 | 33.74 | 42.78 | 31.48 | 30.71 | 40.92 | 50.02 | 34.52 | 42.33 | 45.51 | 49.49 | 37.44 | 33.84 | 28.44 | 31.56 |
|
|
||||||||||||||||
| Taenia solium (NC_004022) | 40.35 | 25.76 | 34.87 | 43.62 | 34.23 | 32.42 | 42.30 | 49.45 | 33.80 | 41.97 | 45.18 | 49.91 | 37.89 | 35.73 | 26.71 | 31.92 |
|
|
||||||||||||||||
| Echinococcus multilocularis (NC_000928) | 43.41 | 25.25 | 37.39 | 43.09 | 34.62 | 33.04 | 39.15 | 44.08 | 36.37 | 44.05 | 47.91 | 48.14 | 38.36 | 35.70 | 29.62 | 33.19 |
|
|
||||||||||||||||
| Echinococcus granulosus (AB786664) | 45.56 | 26.41 | 36.50 | 42.99 | 36.99 | 34.44 | 43.33 | 42.06 | 37.64 | 44.63 | 50.13 | 53.77 | 39.94 | 35.92 | 32.34 | 34.52 |
|
|
||||||||||||||||
| Spirometra erinaceieuropaei (KJ599680) | 44.46 | 32.70 | 45.56 | 48.79 | 42.09 | 34.43 | 51.00 | 38.41 | 41.72 | 49.03 | 53.38 | 57.90 | 43.39 | 41.30 | 31.17 | 37.07 |
|
|
||||||||||||||||
| Spirometra theileri (NC_056327) | 51.92 | 32.70 | 43.95 | 55.45 | 39.35 | 35.52 | 45.32 | 38.41 | 41.60 | 48.11 | 53.51 | 51.59 | 43.89 | 41.67 | 30.96 | 37.19 |
|
|
||||||||||||||||
| Spirometra decipiens (KJ599679) | 44.45 | 32.56 | 41.70 | 50.45 | 41.87 | 35.50 | 47.47 | 36.99 | 43.76 | 49.19 | 51.28 | 51.94 | 43.39 | 43.54 | 31.15 | 38.33 |
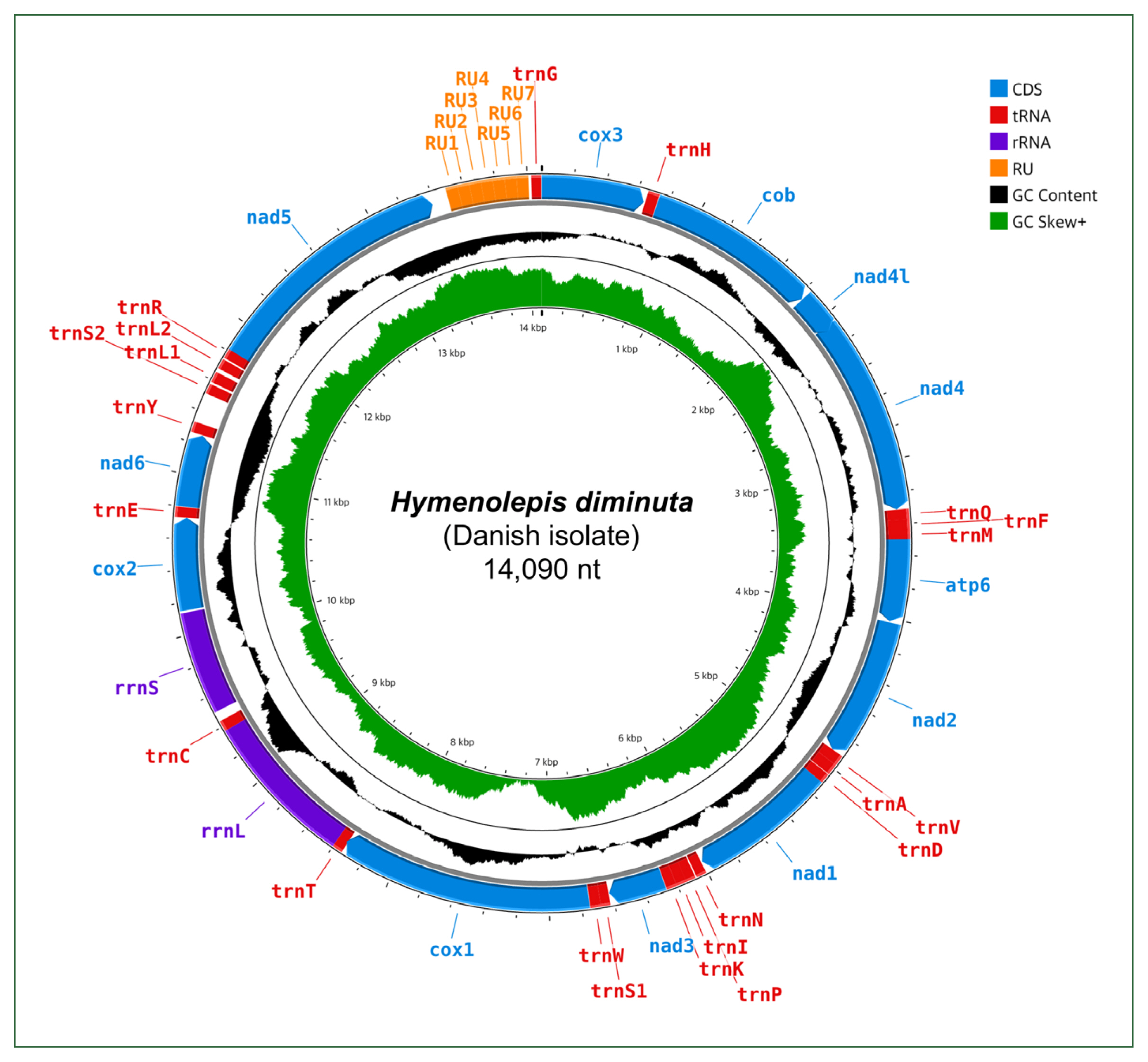
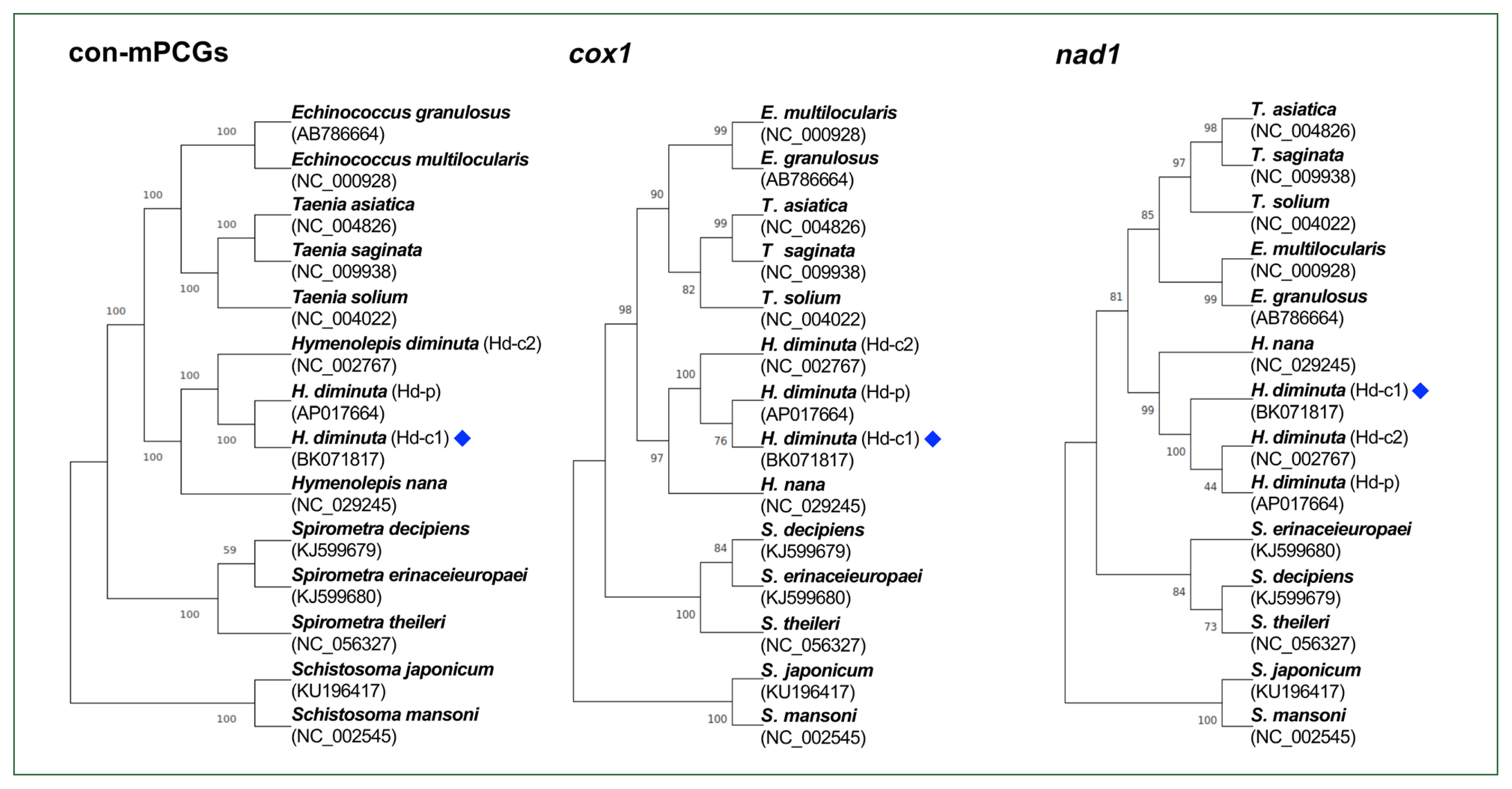
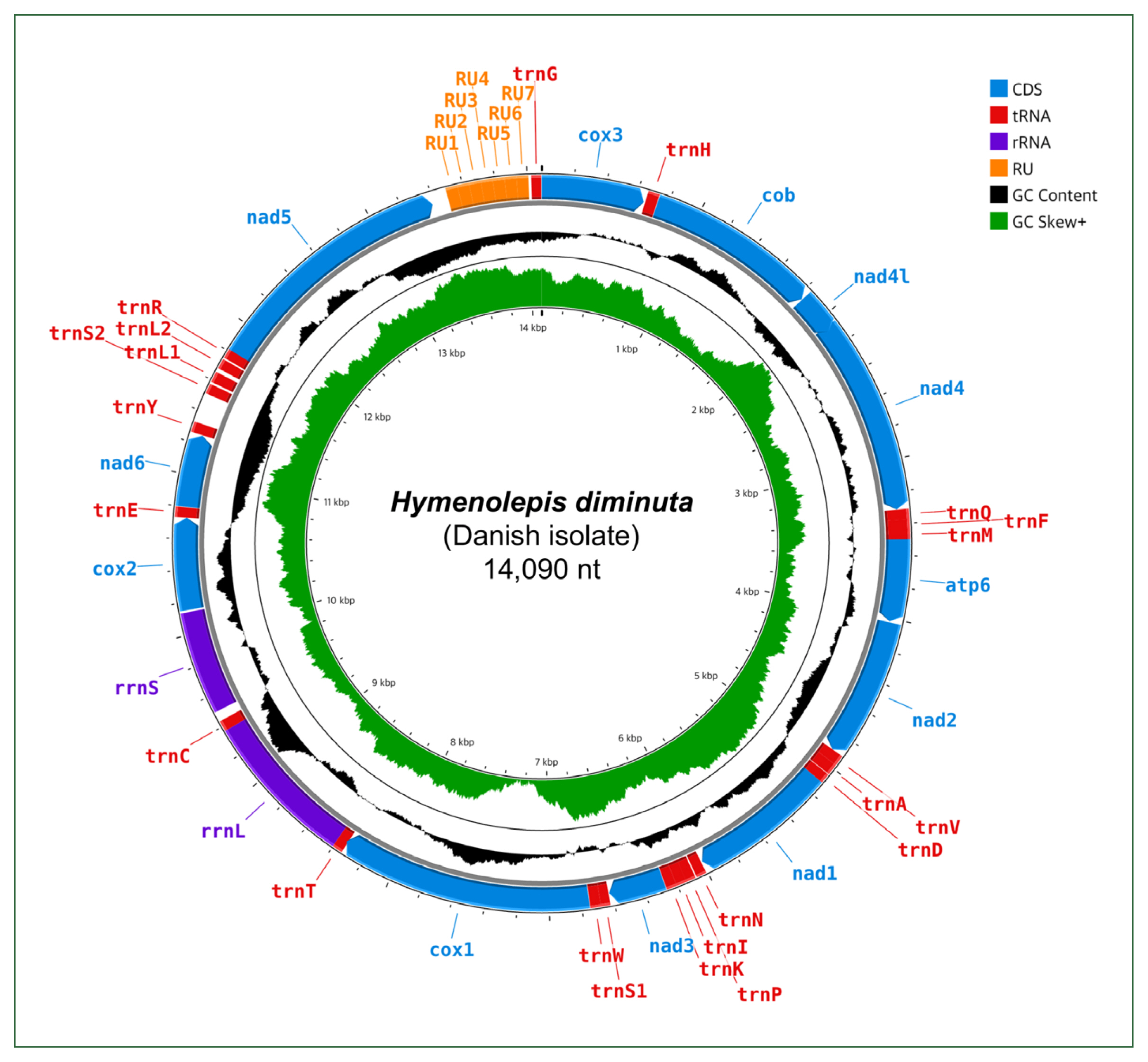
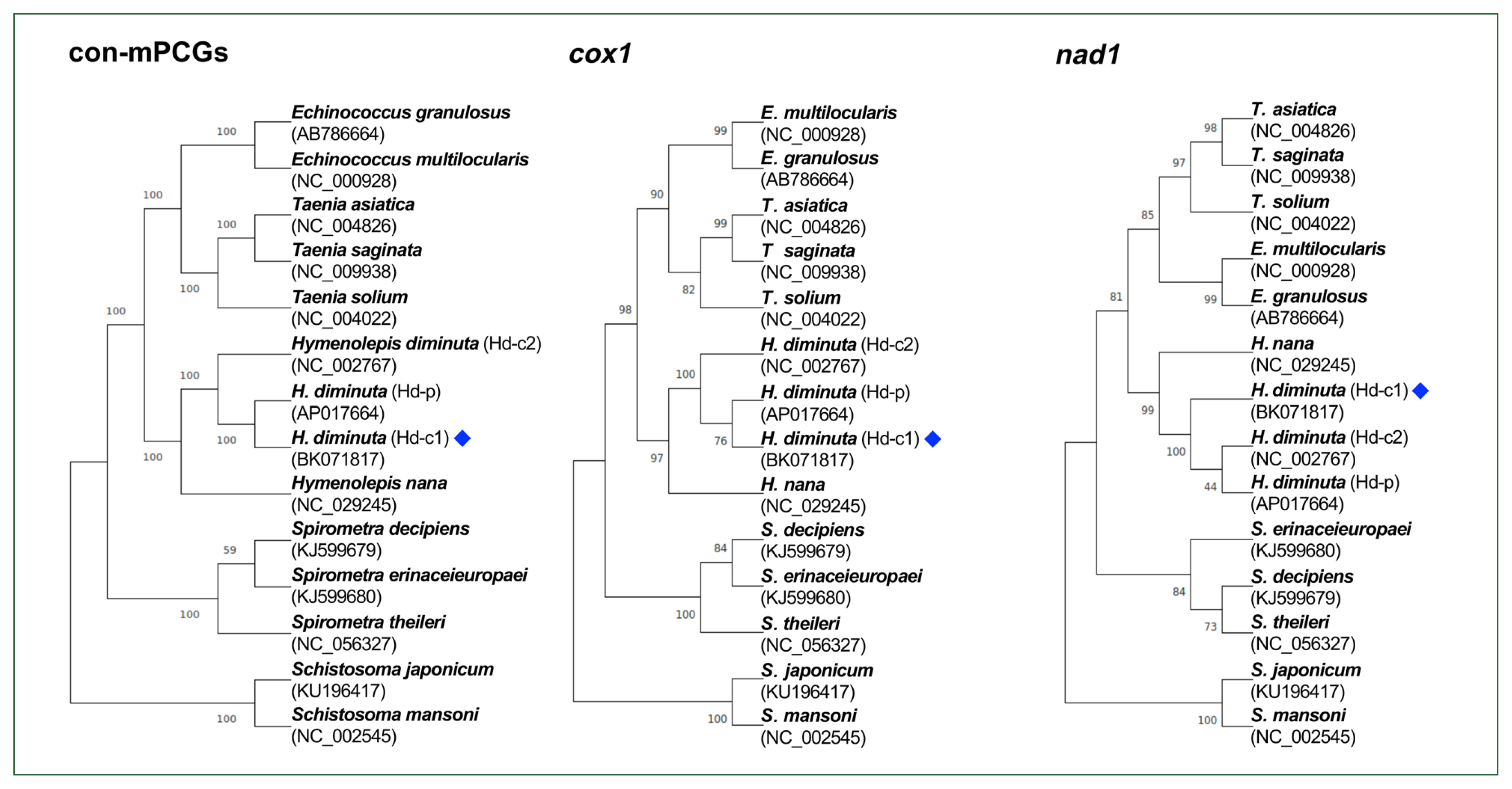
Gene content, length, initiation and stop codons of Hymenolepis diminuta mitogenomes from different datasets, including 2 complete mitogenomes (Hd-c1, BK071817 and Hd-c2, NC_002767) and 1 partial mitogenome (Hd-p, AP017664)
| Gene/region | Positions and size (bp) | Initiation and termination codons | Anticodon | ||||
|---|---|---|---|---|---|---|---|
|
|
|
| |||||
| Hd-c1a | Hd-p | Hd-c2 | Hd-c1 | Hd-p | Hd-c2 | Hd-c1 | |
| cox3 | 1–651 (651) | 130–780 (651) | 7,122–7,772 (651) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
| |||||||
| tRNA-His (H) | 660–731 (72) | 789–860 (72) | 7,781–7,852 (72) | GTG | |||
|
| |||||||
| cytb | 735–1,832 (1,098) | 864–1,961 (1,098) | 7,856–8,953 (1,098) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
| |||||||
| nad4L | 1,836–2,096 (261) | 1,965–2,225 (261) | 8,957–9,217 (261) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
| |||||||
| nad4 | 2,063–3,310 (1,248) | 2,192–3,439 (1,248) | 9,202–10,431 (1,230) | ATG/TAG | ATG/TAG | ATT/TAG | |
|
| |||||||
| tRNA-Gln (Q) | 3,312–3,377 (66) | 3,441–3,505 (65) | 10,433–10,498 (66) | TTG | |||
|
| |||||||
| tRNA-Phe (F) | 3,377–3,439 (63) | 3,506–3,568 (62) | 10,498–10,560 (63) | GAA | |||
|
| |||||||
| tRNA-Met (M) | 3,436–3,499 (64) | 3,565–3,628 (64) | 10,577–10,620 (64) | CAT | |||
|
| |||||||
| atp6 | 3,468–4,019 (552) | 3,633–4,148 (516) | 10,625–11,140 (516) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
| |||||||
| nad2 | 4,026–4,907 (882) | 4,155–5,036 (882) | 11,147–12,028 (882) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
| |||||||
| tRNA-Val (V) | 4,908–4,973 (66) | 5,037–5,102 (66) | 12,029–12,093 (65) | TAC | |||
|
| |||||||
| tRNA-Ala (A) | 4,974–5,043 (70) | 5,103–5,172 (70) | 12,095–12,164 (70) | TGC | |||
|
| |||||||
| tRNA-Asp (D) | 5,048–5,109 (62) | 5,177–5,238 (62) | 12,169–12,230 (62) | GTC | |||
|
| |||||||
| nad1 | 5,110–6,000 (891) | 5,239–6,129 (891) | 12,231–13,121 (891) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
| |||||||
| tRNA-Asn (N) | 6,009–6,072 (64) | 6,138–6,201 (64) | 13,130–13,193 (64) | GTT | |||
|
| |||||||
| tRNA-Pro (P) | 6,081–6,142 (62) | 6,210–6,271 (62) | 13,202–13,264 (63) | TGG | |||
|
| |||||||
| tRNA-Ile (I) | 6,142–6,205 (64) | 6,271–6,334 (64) | 13,264–13,325 (62) | GAT | |||
|
| |||||||
| tRNA-Lys (K) | 6,206–6,269 (64) | 6,335–6,398 (64) | 13,327–13,390 (64) | CTT | |||
|
| |||||||
| nad3 | 6,273–6,620 (348) | 6,402–6,749 (348) | 13,394–13,741 (348) | ATG/TAG | ATG/TAG | ATG/TAG | |
|
| |||||||
| tRNA-Ser (S1) | 6,626–6,684 (59) | 6,755–6,813 (59) | 13,747–13,805 (59) | GCT | |||
|
| |||||||
| tRNA-Trp (W) | 6,685–6,750 (66) | 6,814–6,879 (66) | 13,806–13871 (66) | TCA | |||
|
| |||||||
| cox1 | 6,754–8,346 (1,593) | 6,883–8,475 (1,593) | 4–1,555 (1,552) | ATG/TAG | ATG/TAG | TTT/ATTb | |
|
| |||||||
| tRNA-Thr (T) | 8,337–8,401 (65) | 8,466–8,530 (65) | 1,557–1,621 (65) | TGT | |||
|
| |||||||
| rrnL | 8,402–9,368 (967) | 8,531–9,497 (967) | 1,622–2,588 (967) | ||||
|
| |||||||
| tRNA-Cys (C) | 9,369–9,435 (67) | 9,498–9,564 (67) | 2,589–2,655 (67) | GCA | |||
|
| |||||||
| rrnS | 9498–10,137 (640) | 9,565–10,279 (715) | 2,656–3,364 (709) | ||||
|
| |||||||
| cox2 | 10,151–10,729 (579) | 10,280–10,858 (579) | 3,371–3,949 (579) | ATG/TAA | ATG/TAA | ATG/TAA | |
|
| |||||||
| tRNA-Glu (E) | 10,730–10,794 (65) | 10,859–10,923 (65) | 3,950–4,014 (65) | ||||
|
| |||||||
| nad6 | 10,798–11,256 (459) | 10,927–11,385 (459) | 4,018–4,476 (459) | ATG/TAA | ATG/TAA | ATG/TAA | GTA |
|
| |||||||
| tRNA-Tyr (Y) | 11,260–11,325 (66) | 11,389–11,454 (66) | 4,480–4,545 (66) | TAG | |||
|
| |||||||
| tRNA-Ser (S2) | 11,511–11,573 (63) | 11,640–11,702 (63) | 4,729–4,795 (67) | NGA | |||
|
| |||||||
| tRNA-Leu (L1) | 11,588–11,655 (68) | 11,717–11,784 (68) | 4,808–4,875 (68) | TAA | |||
|
| |||||||
| tRNA-Leu (L2) | 11,681–11,743 (63) | 11,810–11,872 (63) | 4,902–4,964 (63) | TCG | |||
|
| |||||||
| tRNA-Arg (R) | 11,754–11,813 (60) | 11,883–11,942 (60) | 4,975–5,034 (60) | ||||
|
| |||||||
| nad5 | 11,808–13,391 (1,584) | 11,946–13,520 (1,575) | 5,038–6,612 (1,575) | ATG/TAG | ATG/TAG | ATG/TAG | TCC |
|
| |||||||
| tRNA-Gly (G) | 14,025–14,087 (63) | 74–134 (61) | 7,056–7,118 (63) | TTC | |||
|
| |||||||
| Non-coding region | 13,088–14,090 (643) | 13,521–73 (261) | 6,612–7,055 (444) | ||||
|
| |||||||
| Repeat unit | 13,495–14,007 (513) | na | 6,612–6,983 (372) | ||||
na, not available.
aHd-c1 mitogenome begins at 1 bp while Hd-p and Hd-c2 mitogenomes start at 130 bp and 7,122 bp, respectively;
bnon-canonical codons.
Estimation of pairwise distances rate (%) of individual and concatenated mPCGs and MRGs between Hymenolepis diminuta and members of the families such as Hymenolepididae, Taeniidae, and Diphyllobothriidae
| Species/strainsa | mPCGs | MRGs | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
| |||||||||||||||
| atp6 | cox1 | cox2 | cox3 | cytb | nad1 | nad2 | nad3 | nad4L | nad4 | nad5 | nad6 | con-mPCGs | rrnL | rrnS | con-MRGs | |
| H. diminuta (AP017664, Hd-p) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
| ||||||||||||||||
| H. diminuta (NC_002767, Hd-c2) | 0.39 | 0.19 | 0.35 | 0.15 | 0.09 | 0.11 | 0.11 | 0.00 | 0.77 | 0.24 | 0.25 | 0.66 | 0.23 | 0.10 | 0.16 | 0.12 |
|
| ||||||||||||||||
| Hymenolepis nana (NC_029245) | 33.07 | 23.99 | 32.61 | 38.50 | 26.62 | 25.61 | 32.95 | 30.84 | 24.77 | 37.81 | 39.82 | 42.16 | 32.05 | 25.59 | 16.44 | 21.80 |
|
| ||||||||||||||||
| Taenia saginata (NC_009938) | 39.03 | 26.79 | 34.89 | 40.32 | 32.51 | 30.62 | 39.73 | 46.18 | 36.46 | 42.18 | 45.34 | 53.04 | 37.38 | 34.88 | 28.03 | 31.99 |
|
| ||||||||||||||||
| Taenia asiatica (NC_004826) | 40.02 | 26.60 | 33.74 | 42.78 | 31.48 | 30.71 | 40.92 | 50.02 | 34.52 | 42.33 | 45.51 | 49.49 | 37.44 | 33.84 | 28.44 | 31.56 |
|
| ||||||||||||||||
| Taenia solium (NC_004022) | 40.35 | 25.76 | 34.87 | 43.62 | 34.23 | 32.42 | 42.30 | 49.45 | 33.80 | 41.97 | 45.18 | 49.91 | 37.89 | 35.73 | 26.71 | 31.92 |
|
| ||||||||||||||||
| Echinococcus multilocularis (NC_000928) | 43.41 | 25.25 | 37.39 | 43.09 | 34.62 | 33.04 | 39.15 | 44.08 | 36.37 | 44.05 | 47.91 | 48.14 | 38.36 | 35.70 | 29.62 | 33.19 |
|
| ||||||||||||||||
| Echinococcus granulosus (AB786664) | 45.56 | 26.41 | 36.50 | 42.99 | 36.99 | 34.44 | 43.33 | 42.06 | 37.64 | 44.63 | 50.13 | 53.77 | 39.94 | 35.92 | 32.34 | 34.52 |
|
| ||||||||||||||||
| Spirometra erinaceieuropaei (KJ599680) | 44.46 | 32.70 | 45.56 | 48.79 | 42.09 | 34.43 | 51.00 | 38.41 | 41.72 | 49.03 | 53.38 | 57.90 | 43.39 | 41.30 | 31.17 | 37.07 |
|
| ||||||||||||||||
| Spirometra theileri (NC_056327) | 51.92 | 32.70 | 43.95 | 55.45 | 39.35 | 35.52 | 45.32 | 38.41 | 41.60 | 48.11 | 53.51 | 51.59 | 43.89 | 41.67 | 30.96 | 37.19 |
|
| ||||||||||||||||
| Spirometra decipiens (KJ599679) | 44.45 | 32.56 | 41.70 | 50.45 | 41.87 | 35.50 | 47.47 | 36.99 | 43.76 | 49.19 | 51.28 | 51.94 | 43.39 | 43.54 | 31.15 | 38.33 |
mPCG, mitochondrial protein-coding gene; MRG, mitoribosomal gene; con-mPCG, concatenation of 12 mPCG; con-MRG, concatenation of 2 MRG.
aH. diminuta (BK071817, Hd-c1) served as the reference for pairwise distance rate calculations.
na, not available.
Hd-c1 mitogenome begins at 1 bp while Hd-p and Hd-c2 mitogenomes start at 130 bp and 7,122 bp, respectively;
non-canonical codons.
mPCG, mitochondrial protein-coding gene; MRG, mitoribosomal gene; con-mPCG, concatenation of 12 mPCG; con-MRG, concatenation of 2 MRG.
H. diminuta (BK071817, Hd-c1) served as the reference for pairwise distance rate calculations.