Abstract
A complete cDNA sequence encoding a pore-forming subunit (Kir6.2) of ATP-senstive potassium channel in the adult worm, Clonorchis sinensis, termed CsKir6.2, was isolated from an adult cDNA library. The cDNA contained a single open-reading frame of 333 amino acids, which has a structural motif (a GFG-motif) of the putative pore-forming loop of the Kir6.2. Peculiarly, the CsKir6.2 shows a lack-sequence structure, which deleted 57 amino acids were deleted from its N-terminus. The predicted amino acid sequence revealed a highly conserved sequence as other known other Kir6.2 subunits. The mRNA was weekly expressed in the adult worm.
-
Key words: Clonorchis sinensis, potassium channels, cloning
Adenosine 5'-triphosphate-sensitive potassium channels (K
ATP channels) are thought to regulate various cellular functions such as secretion as well as muscular and neural excitability by linking the cell's metabolic state to its membrane potential (
Noma, 1983;
Bernardi et al., 1993). K
ATP channels were originally discovered in the heart (
Noma, 1983) and found later in various tissues, including pancreatic-cells (
Rorsman et al., 1985), smooth and skeletal muscles (
Standen et al., 1989), the brain (
Ashford et al., 1988), pituitary glands (
Bernardi et al., 1988), and the kidney (Hunter et al., 1988). It has been discovered that the K
ATP channels are comprised of two kinds of subunits: sulphonylurea receptors (SUR) which belong to the ATP-binding cassette family and inwardly-rectifying K
+ channel (
Kir) subunits which forms the potassium ion-selective pore. The channels are proposed to form a complex of
SUR1 and
Kir6.2 in pancreatic cells (
Sakura et al., 1995;
Inagaki et al., 1996), and
SUR2A and
Kir6.2 in cardiac and skeletal muscles (
Inagaki et al., 1996). In particular, several
Kir6.2 genes have been identified in human, mouse, rat and rabbit tissues, showing high mRNA levels in the heart, skeletal muscle, bladder and gut. Therefore, the K
ATP channel of the helminth worm is thought to be important in the cellular communication and metabolic pathway. However, the information on the study of the K
ATP channel for the helminth worm has been scare until now.
To add useful information for ion channel of the helminth worm, the gene encoding a pore-forming subunit of the liver fluke in the adult worm Clonorchis sinensis (CsKir6.2) was isolated and characterized. In addition, the gene expression level of a CsKir6.2 in the adult worm was investigated using a Northern blot analysis.
The total RNA was isolated from the adult of
C. sinensis by guanidium isothiocyanate lysis and CsCl centrifugation at 32,000 × g for 16 hr at 20℃. A cDNA library was constructed using SMART cDNA library synthesis kit (Clontech., Palo Alto, CA, USA). Briefly, the first-strand cDNAs were synthesized using PowerScript reverse transcriptase, SMART IV oligonucleotide and a modified oligo (dT) primer (CDS III/3' PCR primer). The second-strand cDNAs were synthesized using 50 × Advantage 2 polylmerase Mix, 5' PCR primer, and a CDS/3' PCR primer. The resulting cDNA was digested with
Sfi I, and fractionated using Chroma Spin-400. The cDNAs larger than 0.5 kb were pooled (data not shown). After digestion with
Sfi I and size fractionation, the cDNA was ligated into the Sfi I-digested λTriplEx2 vector using T4 DNA ligase. The ligated cDNAs were packaged in Gigapack III gold packaging extract (Stratagene, La Jolla, CA, USA). To obtain a partial fragment of a specific probe, 1 gm of total RNA from the adult worm
C. sinensis was reverse-transcribed using an oligo d(T)
15 primer and Moloney Murine Leukemia Virus (M-MuLV) reverse transcriptase (Life Technologies, Gaithersburg, MD, USA). Two degenerate oligonucleo-tides (Kir6.2F, 5-TGACCT GGCSCCCRGCGA-3 and Kir6.2R, 5-CRTCKGC CAGGTAGGAGGT-3 S, G+C; R, A+G; K, G+T) were designed and synthesized according to the highly conserved regions of various human and rabbit ATP-sensitive inwardly-rectifying potassium channels, termed Kir6.2. The resulting cDNA was subjected to PCR amplification in a 50 µl reaction mixture containing 5 µl of RT-PCR product, 5 µl of 10 ×
Taq polymerase buffer, 4 µl of 2.5 mM dNTPs, 25 pM of each primers and 5 units of
Taq polymerase. The reaction was subjected to 35 cycles of denaturation at 95℃ for 30 sec, annealing at 55℃ for 1 min, and extension at 72℃ for 30 sec, and one at 72℃ for 5 min in a thermal cycler (GeneAmp PCR system 9700, Perkin-Elmer). PCR with these primers and the first strand cDNA from the total RNA amplified an approximately 620 bp cDNA fragment, which then was sequenced by direct sequencing. The PCR fragment was showed the significant sequence homology with Kir 6.2 family (data not shown). An adult worm
C. sinensis cDNA library of approximately 100,000 independent plaques was screened by plaque hybridization employing 620 bp partial fragment, which was amplified by RT-PCR. Hybridization to the
32P-labeled probe was done overnight at 42℃. The membrane was washed with high stringency and exposed to Kodak X-AR film. Positive clones were isolated and the lambda DNA was purified by use of a Qiagen Lambda kit (Qiagen, Valencia, CA). The cDNA insert was cloned into a pGEM-T easy vector (Promega) by PCR, using λTriplEx Sequencing primer set (Clontech) and advanced Taq polymerase with proofreading capacity (Clontech). The recombinant plasmid in the bacterial culture was purified with a Qiagen plasmid minikit (Qiagen). The nucleotide sequence was determined by the Dideoxy Chain Termination method using a Sequenase kit (ABI Prism Dye Terminator Cycle Sequencing Core Kit, Perkin Elmer) and an automated DNA sequencer (Applied Biosystems model 377A; Perkin Elmer). The determined nucleotide sequence and deduced amino acid sequence were analyzed with Basic Local Alignment Search Tool (BLAST) in National Center for Biotechnology Information (NCBI) and ExPASy Molecular Biology Server (
http://www.expasy.ch)
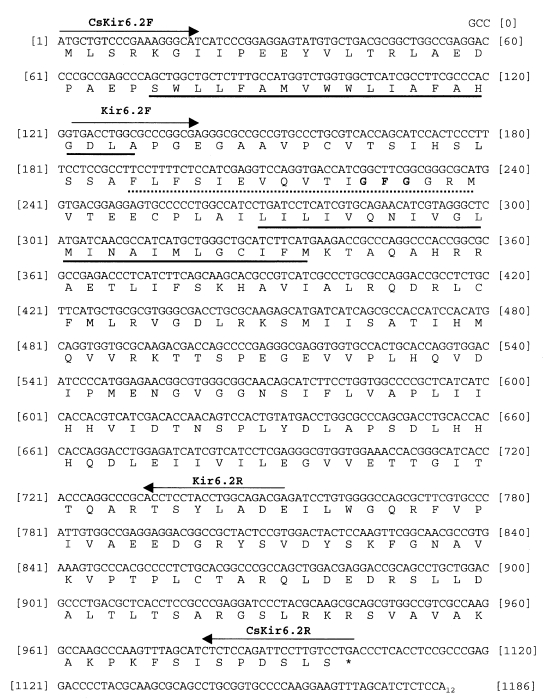
The one clone containing the largest insert (1,189 bp in length) revealed a 999 bp complete open reading frame (ORF) coding for a protein of 333 amino acids. The 3'-untranslated region was contained poly (A)
+ tail (
Fig. 1). This sequence has been deposited in the GenBank database with the accession number AY277926. A hydrophobicity plot of the Kir6.2 revealed two putative transmembrane domains as found in other Kir channels. In addition, a GFG-motif was found in the putative pore-forming loop. The GFG-motif was showed in only both Kir6.1 and Kir6.2 (
Inagaki et al., 1996). Although Kir6.2 shows a high degree of homology with Kir6.1, the intracellular N-terminal and C-terminal regions and an extracellular region between the first transmembrane region and the pore region (H5) do not. Therefore, the nucleotide sequence of CsKir6.2 is more similar to other known Kir6.2 than to other ion channel subunits. However, the CsKir6.2 has more unusual structural features than do other Kir6.2 genes (
Fig. 2). The CsKir6.2 is smaller than Kir6.2 subunits of other species, containing a lacked 57 amino acids sequence in the N-terminal region.
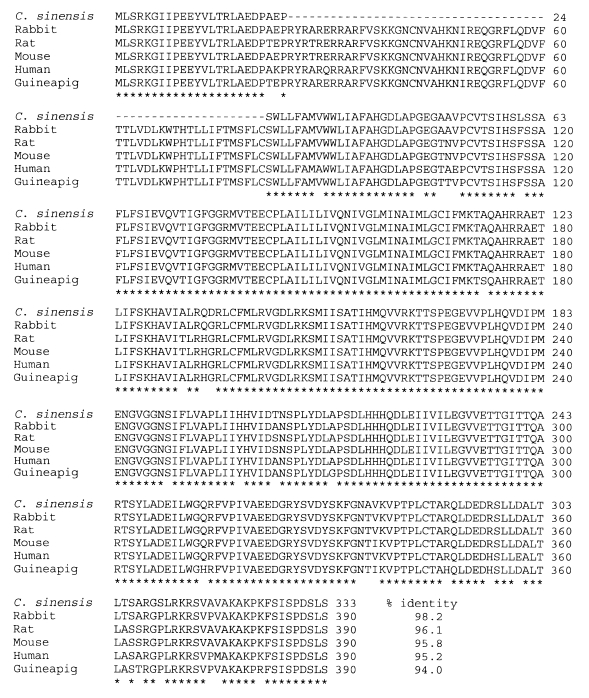
In the case of ruling out a lacked sequence region, a comparison of the amino acid sequence for CsKir6.2 with the Kir6.2 subunits of other species in the database showed a high identity to the proteins of human, mouse, rabbit, guinea pig and rat (
Fig. 2). There was a difference of 13, 14, 6, 20, and 13 amino acids between the sequences of
C. sinensis and the guinea pig, human, mouse, rat, and rabbit, respectively. A Northern blot was used to investigate the expression of Kir6.2 in
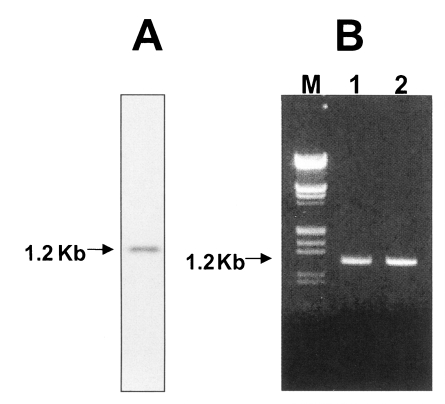
C. sinensis. A single RNA transcript of approximately 1 kb in size were weakly expressed (
Fig. 3A). In order to reveal the cause of a lacked-sequence structure in the Cskir6.2, it was amplified against genomic DNA using CsKir6.2F (5'- ATGCTGTCCCGAAAGGGCAT-3') and CsKir6.2R (5'-TCAGGACAAGGAATCTGGAG-3') primers. Samples were denatured at 95℃ for 2 min, followed by 35 cycles of 94℃ for 30 sec, 58℃ for 30 sec and 72.5℃ for 1 min. A 5 min final extension was performed at 72.5℃. PCR products were analyzed on 1% agarose gel. The nucleotide sequence was determined by dideoxynucleotide chain termination method using a sequenase kit (ABI PRISM Dye Terminator Cycle Sequencing Core Kit) and an automated DNA sequencer. Amplicon of PCR was identical to the cDNA in size (
Fig. 3B) and nucleotide sequence (data not shown). Therefore, the lacked-sequence structure of CsKir6.2 is thought to be a peculiar characteristic of
C. sinensis and is not isoform.
In this study, a pore-forming subunit of the KATP channel (Kir6.2 gene) of C. sinensis was cloned. This is the first report on the cloning of the Kir6.2 gene from trematodes. These results demonstrate that Kir6.2 encodes an inwardly-rectifying KATP channel protein which is expressed in adult C. sinensis. Generally, the KATP channel consists of a pore forming subunit (Kir6.2) and a sulphonylurea receptor (SUR). Both subunits are required to form a functional KATP channel. Therefore, further study is needed to address the exact functions of KATP channel by co-expressing Kir6.2 and SUR after the SUR gene is cloned in C. sinensis.
Notes
-
This work was supported by the 2002 Inje University research grant.
References
- 1. Ashford ML, Sturgess NC, Trout NJ, Gardner NJ, Hales CN. Adenosine-5'-triphosphate-sensitive ion channels in neonatal rat cultured central neurons. Eur J Physiol 1988;412:297-304.
- 2. Bernardi H, Fosset M, Lazdunski M. Characterization, purification, and affinity labeling of the brain [3H]glibenclamide-binding protein, a putative neuronal ATP-regulated K+ channel. Proc Nat Acad Sci USA 1988;85:9816-9820.
- 3. Bernardi H, De-Weille JR, Epelbaum J, et al. ATP-modulated K+ channels sensitive to antidiabetic sulfonylureas are present in adenohypophysis and are involved in growth hormone release. Proc Nat Acad Sci USA 1993;90:1340-1344.
- 4. Inagaki N, Gonoi T, Clement JP, et al. A family of sulfonylurea receptors determines the pharmacological properties of ATP-sensitive K+ channels. Neuron 1996;16:1011-1017.
- 5. Noma A. ATP-regulated K+ channels in cardiac muscle. Nature 1983;305:147-148.
- 6. Rorsman P, Trube G. Glucose dependent K+ channels in pancreatic-cells are regulated by intracellular ATP. Eur J Physiol 1985;405:305-309.
- 7. Sakura H, Ammala C, Smith PA, Gribble FM, Aschcroft FM. Cloning and functional expression of the cDNA encoding a novel ATP-sensitive potassium channel subunit expressed in pancreatic-cells, brain, heart and skeletal muscle. FEBS Lett 1995;377:338-344.
- 8. Standen NB, Quayle JM, Davies NW, Brayden JE, Huang Y, Nelson MT. Hyperpolarizing vasodilators activate ATP-sensitive K+ channels in arterial smooth muscle. Science 1989;245:177-180.
Fig. 1Nucleotide and deduced amino acid sequences of a pore-forming subunit (Kir6.2) of KATP channel from Clonorchis sinensis. The single underline shows the region corresponding to the putative transmembrane, and the dotted underline shows pore domains. An asterisk indicates a stop codon. The bold letter shows a GFG-motif (the putative pore-forming loop). The position of primers for cDNA PCR (Kir6.2F and Kir6.2R) and gDNA PCR (CsKir6.2F and CsKir6.2R) indicated above the nucleotide sequence.

Fig. 2Comparison of deduced amino acid sequences of Clonorchis sinensis Kir6.2 (CsKir6.2), rabbit (Oryctolagus cuniculus, the Genbank No. D45025), rat (Rattus norvegicus, the Genbank No. BAA96239), mouse (Mus musculus, the Genbank No. Q61743), human (Homo spiens, the Genbank accession number Q14654), and guinea pig (Cavia porcellus, the Genbank No. Q9JHJ9). Gaps indicated by dashes (- - -) are introduced to optimize the alignment. An asterisk indicates an amino acid that all peptides show identical one to the query sequence.

Fig. 3Northern blot analysis and genomic DNA amplification of CsKir6.2. (A) The total RNA from adult Clonorchis sinensis was separated on a 1.2% formaldehyde agarose gel and transferred to a nylon membrane. The labeling of the probe and detection of hybridization signal were carried out with a 32P-labelled system. (B) The CsKir6.2 was amplified against the genomic DNA and cDNA of adult C. sinensis, then separated on a 1% agarose gel. Lane M, γDNA/EcoR I + Hind III marker (MBI fermantas); lane 1, PCR-amplified genomic DNA; lane 2, PCR amplified cDNA.

Citations
Citations to this article as recorded by

- Pipeline for the identification and classification of ion channels in parasitic flatworms
Bahiyah Nor, Neil D. Young, Pasi K. Korhonen, Ross S. Hall, Patrick Tan, Andrew Lonie, Robin B. Gasser
Parasites & Vectors.2016;[Epub] CrossRef - Progress on the transcriptomics of carcinogenic liver flukes of humans—Unique biological and biotechnological prospects
Neil D. Young, Aaron R. Jex, Cinzia Cantacessi, Bronwyn E. Campbell, Thewarach Laha, Woon-Mok Sohn, Banchob Sripa, Alex Loukas, Paul J. Brindley, Robin B. Gasser
Biotechnology Advances.2010; 28(6): 859. CrossRef - Functional Genes and Proteins of Clonorchis sinensis
Tae Im Kim, Byoung-Kuk Na, Sung-Jong Hong
The Korean Journal of Parasitology.2009; 47(Suppl): S59. CrossRef - Clonorchiasis: an update
H.-J. Rim
Journal of Helminthology.2005; 79(3): 269. CrossRef