Abstract
This study aimed to identify the assemblages (or subassemblages) of Giardia duodenalis by using normal or nested PCR based on 4 genetic loci: glutamate dehydrogenase (gdh), triose phosphate isomerase (tpi), β-giardin (bg), and small subunit ribosomal DNA (18S rRNA) genes. For this work, a total of 216 dogs' fecal samples were collected in Guangdong, China. The phylogenetic trees were constructed with MEGA5.2 by using the neighbor-joining method. Results showed that 9.7% (21/216) samples were found to be positive; moreover, 10 samples were single infection (7 isolates assemblage A, 2 isolates assemblage C, and 1 isolate assemblage D) and 11 samples were mixed infections where assemblage A was predominant, which was potentially zoonotic. These findings showed that most of the dogs in Guangdong were infected or mixed-infected with assemblage A, and multi-locus sequence typing could be the best selection for the genotype analysis of dog-derived Giardia isolates.
-
Key words: Giardia duodenalis, dog, multi-locus sequence typing, Guangdong, China
Giardia duodenalis is one of the most common intestinal protozoan parasite, which is ubiquitous in mammals [
1]. In Asia, Africa, and Latin America, approximately 200 million people present symptomatic giardiasis and 500,000 new cases are diagnosed per year [
2].
G. duodenalis is considered as a species complex, comprising at least 8 distinct genetic groups, referred to as assemblages A to H [
3]. To date, only assemblages A (subtypes I and II) and B (subtypes I and IV) have been associated with human infections, but are also found in a number of other mammalian hosts [
4]. Genetic assemblages C, D, E, F, G, and H appear to be host-restricted to domestic animals and wild animals [
5]; furthermore, assemblages C and D infect dogs and wild carnivores, assemblage E infects cattle and other hoofed livestock, assemblage F infects cats, assemblage G infects rats [
6], and assemblage H infects marine mammals [
7].
In China, there are approximately 28.5 million cases per year. The majority of giardiasis cases remain unreported [
8]. However, Yong et al. [
9] have obtained 8
G. duodenalis isolates from Anhui Province, China (all from purified cysts); 4 isolates were grouped into assemblage A and 4 into assemblage B. In Henan Province, China, 18
Giardia-positive specimens were characterized by sequence analysis of the triose phosphate isomerase (tpi) gene into assemblages A (8 belonging to AI and 4 AII) and B (belonging to 6 new subtypes) found in 12 and 6 specimens, respectively [
10]. What's more, 5 of 23 isolates from dogs were characterized as assemblage AI and 18 as assemblage D [
11]. Dogs have been considered as the main potential reservoirs because of their close contacts with humans, but direct evidence for animal-to-human transmission is still scarce [
1,
12]. So far, most studies have been mainly based on analysis of the small subunit ribosomal locus (18S rRNA), β-giardin (bg), glutamate dehydrogenase (gdh), elongation factor 1-alpha (ef-1), and triose phosphate isomerase (tpi). Only bg, gdh, and tpi genes were sufficiently discriminatory to type at the level of subassemblage, 18S rRNA and ef-1 permit identification only at the level of the assemblage, which are much more conserved than other three loci [
6,
13].
To date, due to the lack of high-resolution genotype identification based on 1 gene, a multi-locus sequence-typing scheme is needed on the molecular identification of zoonotic assemblages in dogs. Therefore, the aim of the present study was to genetically characterize isolates of G. duodenalis from dogs in Guangdong, China using analysis of tpi, gdh, bg, and 18S rRNA loci combined.
A total of 216 dog fecal samples were obtained from 4 different shelters in Guangdong, China during June 2012 to October 2013. Two shelters were located in Shunde (urban) and Sanshui (suburban) districts of Foshan City, Guangdong and 2 in Baiyun (urban) and Conghua (suburban) districts of Guangzhou City, Guangdong. Thirty fecal samples were collected in Shunde district, 55 in Sanshui district, 80 in Conghua district, and 51 in Baiyun district. All the dogs were abandoned or stray dogs and had stayed in the shelters for at least 1 month. Fresh fecal samples from each dog were transported back to the Parasitology and Parasitic Diseases Laboratory, College of Veterinary Medicine, South China Agricultural University (SCAU) on the same day of collection, preserved in 2.5% potassium dichromate and kept at 4℃. Direct microscopic examination was done after centrifugation of 1.5 g feces by the flotation technique with saturated glucose.
Positive samples examined by microscopy were pretreated with 5 cycles of heating at 100℃ for 5 min and immediate freezing at -80℃ for 5 min. DNA was extracted directly from fecal samples using a commercial DNA extraction kit (QIAamp DNA Stool Mini Kit, QIAgen, Hilden, Germany) according to the manufacturer's instructions. DNA samples were stored at -20℃ until use.
The 18S rRNA, bg, and gdh genes were amplified individually according to the protocols described previously [
6,
14,
15]. The tpi primers, tpi1 (5' AACGCAATCACTGTATCT 3') and tpi2 (5' CAATGACAACCTCCTTCC 3') were used for the primary amplification, and tpi3 (5' CTTCATCGGCGGTAACTT 3') and tpi4 (5' GGCACGCTTAGCCTTCTT 3') were used for the nested amplification. Identical conditions for the primary and secondary amplification of the tpi gene fragment were as following: 35 cycles (94℃ for 30 sec, 45℃ for 30 sec, and 72℃ for 45 sec) in a T-personal thermocycler, with an initial preheating at 94℃ for 5 min and a final extension at 72℃ for 7 min. PCR products were analyzed after electrophoresis in 1.5% agarose gels and stained with 0.2 g/ml of ethidium bromide and visualized on a UV transilluminator. Positive amplicons were purified and sequenced at an ABI 377 automated DNA sequencer (BigDye Terminator Chemistry). Obtained sequences were aligned by Clustal X (2.21) using default parameters, and analyzed by MEGA5.2 software. Obtained nucleotide sequences have been submitted to the GenBank™, under accession numbers (KJ027399 to KJ027403 and KJ027407) for 18S rRNA; (KJ027417 to KJ027423, KJ027411, KJ027413, KJ0274134, KJ027415, and KJ027425) for bg, (KJ027426 to KJ027433, and KJ027438) for gdh, and (KJ027448 to KJ027463) for tpi.
A 334 bp fragment of the tpi gene was obtained from 16 dog's isolates; the sequence analysis revealed assemblage AI in 15 isolates and assemblage D in 1 isolate. The amplification of a 515 bp fragment of the bg gene was obtained from 12 dog's isolates; sequence analysis revealed assemblage AI in 1 isolate, assemblage C in 3 isolates, and assemblage D in 8 isolates. The amplification of a 530 bp fragment of the gdh gene was obtained from 8 dog isolates; sequence analysis revealed assemblage AI in 1 isolate, assemblage C in 3 isolates, and assemblage D in 4 isolates. The amplification of a 292 or 291 bp fragment of the 18S rRNA gene was obtained from 6
G. duodenalis isolates, the sequence analysis revealed assemblage A in 4 isolates, assemblage C in 1 isolate, and assemblage D in 1 isolate (
Table 1).
A single infection with assemblage A was found in 7 samples, where 1 was typed by 3 loci, 3 were typed by 2 loci, and 3 was typed by 1 locus. A single infection with assemblage C was found in 2 samples, where 1 was typed by 2 loci and 1 was typed by 1 locus. A single infection with assemblage D was found in 1 sample that was typed at 3 loci. Among mixed infections, 2 assemblages were detected in 11 samples (A and D in 7, D and C in 2, A and C in 2), more than 2 assemblages in the same sample were not found (
Table 2).
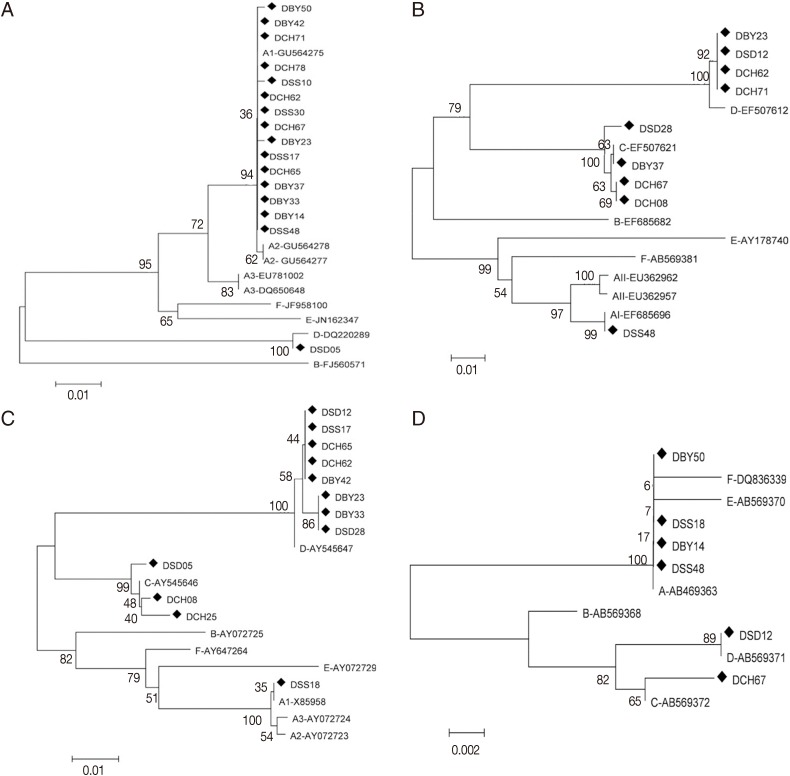
The phylogenetic analysis was performed by using the neighbor-joining method based on 4 gene loci. Result showed that assemblages A and D were significantly separated at the tpi gene. Assemblage A subgroups remained separated in the tpi gene. All sequences of assemblage A were placed in 1 cluster with AI sequence reference (GU564275) separating from AII and AIII in the neighbor joining tree (
Fig. 1A). Assemblages A, C, and D were significantly separated at the gdh gene, and subgrouping was observed among assemblages A in the gdh analysis. The neighbor-joining tree placed the representative sequence (DSS48) in 1 cluster with AI sequence reference (EF685696) with high bootstrap support (
Fig.1B). Assemblages A, C, and D were significantly separated at the bg (
Fig. 1C) and 18S rRNA genes (
Fig. 1D). At bg gene, the sequence (DSS18) were placed in 1 cluster with AI sequence reference (AB469365) separating from AII and AIII. Phylogenetic tree of the tpi, gdh, bg, and 18S rRNA genes by the maximum-likelihood method were similar to that by the neighbor-joining method.
The homologous analysis was performed by using nucleotide sequence based on tpi, gdh, and bg gene loci. At tpi gene, all sequences of assemblage A showed complete homology to AI except for DSS10 (G-A at position 50), DBY50 (A-G at position 146) and DBY23 (T-C at position 246) of dog isolates. At gdh gene, DSS48 sequence showed complete homology to the AI reference sequence (EF685696). At bg gene, DSS18 sequence showed complete homology to AI reference sequence (AB469365) (
Table 3).
The current study present the first genotyping by multi-locus sequence typing of
G. duodenalis isolated from dogs in Guangdong, China. Most of the dogs in this study were infected or mixed-infected with assemblage A, which is different from most previous studies. Recently, Feng and Xiao [
8] have stated that 1,049 of 1,563 (67%)
G. duodenalis isolates from dogs were either assemblage C or D in the world. This phenomenon may be related to the living environments of the dogs, as they lived in shelters, the first infection by assemblage A may predominate over the shelters latterly [
13]. On the other hand, dogs from shelters which crowded breeding show higher rate of dog-to-dog transmission and so do the young animals than older ones [
16]. Epe et al. [
17] have reported an average rate of 24.8% in dogs through Europe, which rises to 34.7% in shelter dogs.
In our study, the tpi locus show the highest amplification rate in dogs, where most of samples were typed as assemblage AI except 1 (DSD05) which was typed as assemblage D. The sensitivity of tpi gene seemed to be high in amplifying assemblage A which has zoonotic potential. Two loci gdh and bg appeared more likely to amplify assemblage C and D in dogs, which is in coherence with Scorza et al. [
18]. Moreover, the gdh and bg gene also amplified 1 assemblage AI in dogs respectively, which have zoonotic potential with certainly. Our results showed the presence of potentially zoonotic assemblages in dogs in Guangdong, China, which is consistent with Li et al. [
11]. Comparing the conserved marker (18S) with variant marker (bg, gdh, and tpi) in genotyping and the amplification rate, the PCR amplification at the 18S rRNA loci had the poorest performance, but with longer fragment (about 292 bp) in this study than most of the previous studies. To date, almost half of the studies performed were based on the analysis of a short fragment (130 bp) of 18S rRNA locus [
19]. Furthermore, studies on
G. duodenalis have carried out for several years in our lab, and no assemblage B was found in dog's isolates from Guangdong, South China. The assemblage B isolates might not exist in this region. Thus, more researches are strongly required to clarify the presence/absence of assemblage B.
The high frequency of multiple/mixed infections found in this study has been increasingly reported in other countries on multi-locus studies in both humans and mammalians including dog, cat, cattle, goat, sheep, pig, and wildlife specimens [
6,
8,
12,
13,
20,
21,
22]. Eleven samples were mixed infections where assemblage A was predominant in the study. It may be attributed to mixed infections or allelic sequence heterozygosity (ASH). Mixed infections can happen when a host ingests
Giardia cysts of different genetic profiles or subsequent infection of an infected host by genetically different
Giardia cysts. This is especially common in areas where giardiasis is endemic [
3,
6,
12,
20,
23], but ASH can also account for this finding and sexual reproduction may influence ASH levels [
12]. However, the contribution between mixed infections and ASH in the complexity of genotyping is currently uncertain.
In conclusion, assemblage A was most frequent among infected dogs, which is potentially zoonotic genotype. Moreover, dog giardiasis seems to be more likely dog-to-dog transmission, but humans may be involved in a zoonotic cycle of transmission, where human-to-human transmission is much more important than animal-to-human transmission. As well, more studies on the contributing factors between mixed infections and ASH should be seriously taken place. The choice of gene loci can influence the amplification results, hence, detection methods targeting loci can be extremely useful when a common source of contamination is certainly involved. Thus, multi-locus sequence typing could be considered for the genotype analysis of dog-derived Giardia isolates.
National Natural Science Foundation of China3127255130972179
Notes
-
We have no conflict of interest related to this work.
ACKNOWLEDGMENTS
This work was supported by grant from National Natural Science Foundation of China (grant nos. 31272551 and 30972179). We thank the humane shelter' personnel for helping us in collecting all of the samples.
References
- 1. Cooper MA, Sterling CR, Gilman RH, Cama V, Ortega Y, Adam RD. Molecular analysis of household transmission of Giardia lamblia in a region of high endemicity in Peru. J Infect Dis 2010;202:1713-1721.
- 2. Thompson RCA, Hopkins RM, Homan WL. Nomenclature and genetic groupings of Giardia infecting mammals. Parasitol Today 2000;16:210-213.
- 3. Monis PT, Caccio SM, Thompson RCA. Variation in Giardia: towards a taxonomic revision of the genus. Trends Parasitol 2009;25:93-100.
- 4. Vanni I, Cacciò SM, van Lith L, Lebbad M, Svärd SG, Pozio E, Tosini F. Detection of Giardia duodenalis assemblages A and B in human feces by simple, assemblage-specific PCR assays. PLoS Negl Trop Dis 2012;6:e1776.
- 5. Bertrand I, Albertini L, Schwartzbrod J. Comparison of two target genes for detection and genotyping of Giardia lamblia in human feces by PCR and PCR-restriction fragment length polymorphism. J Clin Microbiol 2005;43:5940-5944.
- 6. Caccio SM, Ryan U. Molecular epidemiology of giardiasis. Mol Biochem Parasitol 2008;160:75-80.
- 7. Lasek-Nesselquist E, Welch DM, Sogin ML. The identification of a new Giardia duodenalis assemblage in marine vertebrates and a preliminary analysis of G. duodenalis population biology in marine systems. Int J Parasitol 2010;40:1063-1074.
- 8. Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev 2011;24:110-140.
- 9. Yong TS, Park SJ, Hwang UW, Yang HW, Lee KW, Min DY, Rim HJ, Wang Y, Zheng F. Genotyping of Giardia lamblia isolates from humans in China and Korea using ribosomal DNA sequences. J Parasitol 2000;86:887-891.
- 10. Wang R, Zhang X, Zhu H, Zhang L, Feng Y, Jian F, Ning C, Qi M, Zhou Y, Fu K, Wang Y, Sun Y, Wang Q, Xiao L. Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp Parasitol 2011;127:42-45.
- 11. Li J, Zhang P, Wang P, Alsarakibi M, Zhu H, Liu Y, Meng X, Li J, Guo J, Li G. Genotype identification and prevalence of Giardia duodenalis in pet dogs of Guangzhou, Southern China. Vet Parasitol 2012;188:368-371.
- 12. Sprong H, Caccio SM, van der Giessen JWB, Network Z. Partners identification of zoonotic genotypes of Giardia duodenalis. PLoS Negl Trop Dis 2009;3:e558.
- 13. Covacin C, Aucoin DP, Elliot A, Thompson RCA. Genotypic characterisation of Giardia from domestic dogs in the USA. Vet Parasitol 2011;177:28-32.
- 14. Hopkins RM, Meloni BP, Groth DM, Wetherall JD, Reynoldson JA, Thompson RCA. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J Parasitol 1997;83:44-51.
- 15. Sulaiman IM, Jiang JL, Singh A, Xiao LH. Distribution of Giardia duodenalis genotypes and subgenotypes in raw urban wastewater in Milwaukee, Wisconsin. Appl Environ Microbiol 2004;70:3776-3780.
- 16. Tangtrongsup S, Scorza V. Update on the diagnosis and management of Giardia spp. infections in dogs and cats. Top Companion Anim Med 2010;25:155-162.
- 17. Epe C, Rehkter G, Schnieder T, Lorentzen L, Kreienbrock L. Giardia in symptomatic dogs and cats in Europe- results of a European study. Vet Parasitol 2010;173:32-38.
- 18. Scorza AV, Ballweber LR, Tangtrongsup S, Panuska C, Lappin MR. Comparisons of mammalian Giardia duodenalis assemblages based on the beta-giardin, glutamate dehydrogenase and triose phosphate isomerase genes. Vet Parasitol 2012;189:182-188.
- 19. Ballweber LR, Xiao L, Bowman DD, Kahn G, Cama VA. Giardiasis in dogs and cats: update on epidemiology and public health significance. Trends Parasitol 2010;26:180-189.
- 20. Huey CS, Mahdy MAK, Al-Mekhlafi HM, Nasr NA, Lim YAL, Mahmud R, Surin J. Multilocus genotyping of Giardia duodenalis in Malaysia. Infect Genet Evol 2013;17:269-276.
- 21. Hussein AIA, Yamaguchi T, Nakamoto K, Iseki M, Tokoro M. Multiple-subgenotype infections of Giardia intestinalis detected in Palestinian clinical cases using a subcloning approach. Parasitol Int 2009;58:258-262.
- 22. Lebbad M, Petersson I, Karlsson L, Botero-Kleiven S, Andersson JO, Svenungsson B, Svard SG. Multilocus genotyping of human Giardia isolates suggests limited zoonotic transmission and association between assemblage B and flatulence in children. PLoS Negl Trop Dis 2011;5:e1262.
- 23. Beck R, Sprong H, Pozio E, Caccio SM. Genotyping Giardia duodenalis isolates from dogs: lessons from a multilocus sequence typing study. Vector Borne Zoonotic Dis 2012;12:206-213.
Fig. 1Phylogenetic relationships of Giardia duodenalis assemblages from dogs inferred by the neighbor-joining analysis of 4 gene loci. (A) Triose phosphate isomerase (tpi) nucleotide sequences. (B) Glutamate dehydrogenase (gdh) nucleotide sequences. (C) β-giardin (bg) nucleotide sequences. (D) Small subunit ribosomal DNA (18S rRNA) nucleotide sequences. Bootstrap values obtained from 1,000 replicates are indicated on branches in percentage.

Table 1.Genotyping data of Giardia intestinalis isolates from dogs in Guangdong, China at the 4 loci tpi, gdh, bg, and 18S rRNA Neg, negative
Table 1.
|
Isolates |
tpi |
gdh |
bg |
18S rRNA |
|
DBY14 |
AI |
Neg |
Neg |
A |
|
DBY23 |
AI |
D |
D |
Neg |
|
DBY33 |
AI |
Neg |
D |
Neg |
|
DBY37 |
AI |
C |
Neg |
Neg |
|
DBY42 |
AI |
Neg |
D |
Neg |
|
DBY50 |
AI |
Neg |
Neg |
A |
|
DCH08 |
Neg |
C |
C |
Neg |
|
DCH25 |
Neg |
Neg |
C |
Neg |
|
DCH62 |
AI |
D |
D |
Neg |
|
DCH65 |
AI |
Neg |
D |
Neg |
|
DCH67 |
AI |
C |
Neg |
C |
|
DCH71 |
AI |
D |
Neg |
Neg |
|
DCH78 |
AI |
Neg |
Neg |
Neg |
|
DSS10 |
AI |
Neg |
Neg |
Neg |
|
DSS17 |
AI |
Neg |
D |
Neg |
|
DSS18 |
Neg |
Neg |
AI |
A |
|
DSS30 |
AI |
Neg |
Neg |
Neg |
|
DSS48 |
AI |
AI |
Neg |
A |
|
DSD05 |
D |
Neg |
C |
Neg |
|
DSD12 |
Neg |
D |
D |
D |
|
DSD28 |
Neg |
C |
D |
Neg |
Table 2.Assemblages and subassemblages of 21 positive samples from dogs at 1, 2, or 3 loci
Table 2.
|
Locus/Loci |
Assemblages and sub-assemblages |
|
1 locus |
tpi |
gdh |
bg |
18S |
|
|
|
AI (3) |
- |
C (1) |
- |
|
|
|
2 loci |
tpi, bg |
tpi, gdh |
tpi, 18S |
bg, gdh |
bg, 18S |
gdh, 18S |
|
AI, D (4) |
AI, C (1) |
AI, A (2) |
C, C (1) |
AI, A (1) |
- |
|
D, C (1) |
AI, D (1) |
|
D, C (1) |
|
|
|
3 loci |
tpi, bg, gdh |
tpi, gdh, 18S |
bg, gdh, 18S |
tpi, bg, 18S |
|
|
|
AI, D, D (2) |
AI, AI, A (1) |
D, D, D (1) |
- |
|
|
|
|
AI, C, C (1) |
|
|
|
|
Table 3.Summary of nucleotide variations of Giardia intestinalis A genotype subassemblages in dog isolates at the bg, tpi, and gdh genes
Table 3.
|
Alignment position |
|
Reference sequences |
Bg |
68 |
317 |
327 |
335 |
473 |
|
|
|
|
|
|
|
|
|
|
|
|
AI-AB469365 |
T |
T |
C |
T |
C |
|
|
|
|
|
|
|
|
|
|
|
|
AII-AY072723 |
C |
C |
. |
. |
T |
|
|
|
|
|
|
|
|
|
|
|
|
AIII-AY072724 |
C |
C |
T |
C |
T |
|
|
|
|
|
|
|
|
|
|
|
|
Dog |
DSS18 |
C |
C |
. |
. |
. |
|
|
|
|
|
|
|
|
|
|
|
|
Reference sequences |
Gdh |
63 |
118 |
162 |
180 |
258 |
312 |
366 |
390 |
420 |
426 |
429 |
453 |
461 |
|
|
|
|
AI-EF685696 |
C |
T |
T |
C |
T |
C |
C |
C |
T |
T |
T |
C |
C |
|
|
|
|
AII-EU362962 |
A |
. |
C |
T |
C |
T |
T |
T |
C |
C |
C |
T |
T |
|
|
|
|
Dog |
DSS48 |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
|
|
|
|
Reference sequences |
Tpi |
25 |
50 |
58 |
73 |
76 |
82 |
85 |
94 |
98 |
109 |
127 |
139 |
146 |
154 |
196 |
246 |
|
AI-GU564275 |
T |
G |
T |
C |
G |
C |
C |
T |
G |
G |
A |
A |
A |
A |
C |
T |
|
AII-GU564277 |
. |
. |
. |
. |
. |
T |
T |
|
T |
A |
G |
G |
. |
G |
T |
. |
|
AIII-EU781002 |
|
|
C |
T |
A |
. |
. |
C |
. |
. |
. |
. |
. |
. |
. |
. |
|
Dog |
DSS10 |
|
A |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
|
DBY50 |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
G |
. |
. |
. |
|
DBY23 |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
C |
|
DSS30/ DSS17/ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
DCH78/ DCH71/ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
DCH67/ DCH65/ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
DCH62/ DBY42/ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
DBY37/ DBY33/ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
DBY14/DSS48 |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |