Abstract
Schistosoma haematobium is one of the most prevalent parasitic flatworms, infecting over 112 million people in Africa. However, little is known about the genetic diversity of natural S. haematobium populations from the human host because of the inaccessible location of adult worms in the host. We used 4 microsatellite loci to genotype individually pooled S. haematobium eggs directly from each patient sampled at 4 endemic locations in Africa. We found that the average allele number of individuals from Mali was significantly higher than that from Nigeria. In addition, no significant difference in allelic composition was detected among the populations within Nigeria; however, the allelic composition was significantly different between Mali and Nigeria populations. This study demonstrated a high level of genetic variability of S. haematobium in the populations from Mali and Nigeria, the 2 major African endemic countries, suggesting that geographical population differentiation may occur in the regions.
-
Key words: Schistosoma haematobium, allelic diversity, allelic composition, microsatellite, Mali, Nigeria
Schistosomiasis is one of the most prevalent parasitic diseases, infecting over 206 millions of people all over the world [
1]. It was estimated that 112 million people were infected with
S. haematobium [
2]. Despite enormous number of people infected with
S. haematobium, empirical studies on genetic diversity of natural
S. haematobium are minimal [
3,
4]. For instance, by using enzyme analyses, 22 laboratory bred isolates of
S. haematobium have shown regional genetic variation [
5]. Moreover, sequence variation has been demonstrated in the complete mitochondrial genome of
S. haematobium, showing population level differences [
6]. More recently, microsatellite markers, which is a powerful tool for genotyping in use today [
7], have been developed for
S. haematobium [
8]. However, little is known about the genetic diversity of natural
S. haematobium populations from the human host.
The inaccessibility of adult
S. haematobium worms, due to their sequestration within the vasculature of the human host, is the main limitation to investigate the genetic diversity of natural
S. haematobium. The laboratory harvest of adult worms involves collection of eggs from the urine of infected individuals and subsequent passage through laboratory populations of snails and rodents [
9]. This laborious approach inevitably introduces a bias, as those parasitic genotypes that were better adapted for laboratory hosts may be artificially selected, and thus could be poor representatives of natural populations [
10].
Pooling of templates has been suggested to reduce the cost of genotyping individuals [
11], especially when inadequate DNA is available from single samples. Pooled DNA samples have been applied to assess allele frequencies in various DNA samples [
12]. In a novel study, pooling DNA was tested for
S. mansoni miracidia by using laboratory isolates and synthetic pools [
13]. It was suggested that pooling is a reliable way to reconstruct genetic features of the population, from which microsatellite allele frequencies can be estimated [
13]. In this study, by using 4 published microsatellite loci, we genotyped individually pooled
S. haematobium eggs of patients' urine samples from 4 locations in the endemic areas of Mali and Nigeria. We aimed to explore the differences in allelic diversity and composition among the populations.
By applying a set of microsatellite markers, we genotyped pooled eggs of
S. haematobium sampled directly from patients’urine samples. The
S. haematobium egg samples were collected from 3 locations in Nigeria (Ebonyi, Bayelsa, and Ogun) and 1 location in Mali (Bamako) (
Fig. 1). The egg samples were obtained by filtering single urine samples of infected primary school students, during May to November 2011. In total, 22 patients from Nigeria and 27 patients from Mali were enrolled for this study; all the patients were confirmed by the presence of
S. haematobium eggs in the urine. The urine samples were then concentrated, and 5 eggs from each patient sample were fixed on Whatman FTA cards. The egg samples were delivered to the National Institute of Parasitic Diseases (NIPD), Chinese Center for Disease Control and Prevention (China CDC) for the study. The study protocol was approved by the Institutional Ethics Committee of NIPD, China CDC. The investigation aim, potential risks, and the benefits were explained to the participants, and the informed consents were obtained verbally. Treatment was followed to the subjects enrolled.
The DNA was extracted from each egg pool by using the DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany), according to the method described by Reinstrup et al. [
14], and stored at 4°C for use. We genotyped each DNA sample using 4 microsatellite markers (A1, A6, B4, and C2) [
8]. All PCRs were performed on a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems, Foster City, California, USA). According to the protocol reported [
8], the PCR products were diluted in autoclaved de-ionized water, and then analyzed on an ABI 3730 capillary automated sequencer, by using a LIZ 500 labelled size standard. Allele sizes were read using GeneMapper version 4.0 (Applied Biosystems). Across different runs of genotyping, the consistency of alleles was examined with 1
Schistosoma genotype as a reference in each run. Alleles at each locus were determined based on the base-pair length of the fragments, and compared with the reference genotype.
For each locus, we applied Student’s t-test to compare the mean allele number of individuals between Mali and Nigeria, with R-software [
15]. In addition, we compared the allelic composition among the populations at location level (Ebonyi, Bayelsa, and Ogun) within Nigeria for each locus, using a r×c test [
16]. As 4 loci were tested, sequential Bonferroni corrections were applied when interpreting the results. Finally, we used a Monte Carlo approach with 10
5 simulation runs [
17], to compare the allelic composition between the Mali and Nigeria samples.
The results indicated that the mean allele number ranged from 2.3 to 5.9 across all loci, and the allele size ranged 110-232 bp at locus A1, 103-364 at A6, 118-365 at B4, and 107-360 at C2 (
Table 1). Secondly, we found that the mean number of alleles per locus in the population from Mali was significantly higher than that from Nigeria at the locus A6 (5.8 vs 4.3 alleles;
t=2.32,
P=0.02), B4 (5.9 vs 2.3;
t=4.76,
P<0.001), and C2 (4.3 vs 2.3;
t=4.38,
P<0.001), except at A1 (5.1 vs 4.7;
t=0.86;
P=0.39) (
Table 2). We did not detect significant differences in allelic composition among the samples from 3 locations in Nigeria at any of the 4 loci, assessed by the Monte Carlo simulation approach (
Fig. 2). Meantime, the results showed that the allelic composition differed significantly between the Mali and Nigeria populations at 3 loci (A6, B4, and C2), but not at A1 (
P=0.25) (
Fig. 3).
The levels of genetic diversity was an important indicator for monitoring the effects of selective pressure imposed by drug treatment and may be a key epidemiologocal component [
18]. The genetic diversity was reported to be unexpectedly low in
S. haematobium, by using DNA barcoding approaches [
19]. However, we found that high allelic diversity of
S. haematobium exists in the populations in Mali and Nigeria, which is consistent with the results previously reported [
20]. It is because the microsatellite marker is particularly powerful to detect nucleotide polymorphisms in
S. haematobium populations [
4]. We also found that the allelic richness of individuals in Mali was higher than that in Nigeria. Referring to the fact that the estimated prevalence of schistosomiasis was 60% in Mali and 23.2% in Nigeria in 2003 [
21], our result supports the view that the higher prevalence of parasite may be closely related to the genetic diversity of parasite populations [
22]. The lower prevalence of schistosomiasis in Nigeria may lead to less gene flow and thus reduce the genetic diversity. It was not surprising that this pattern was consistent with the most recent findings that the
S. haematobium population from Zanzibar having a high prevalence of schistosomiasis possesses larger number of alleles, whereas the lower number of alleles occurred in the South Africa with a low prevalence [
21,
23]. However, the genetic diversity in terms of average allele number in this study did not show significant difference between Ebonyi and Bayelsa samples from Nigeria (excluding the Ogun sample which was collected from only 1 patient; data not shown). The explanation would be that Ebonyi and Bayelsa are closely located, thus a potential gene flow would be expected between.
Previous studies have suggested that human movements within a country may lead to significant gene flow between
S. mansoni populations [
20]. Since both Ebonyi and Ogun are in forest zones, more human movements and water contacts would occur, comparing with Mali, which is largely in the Sahara desert. Therefore, the higher gene flow among the
Schistosoma populations in Nigeria might reduce the opportunities of local adaptations, leading to a lower population differentiation among the populations. When comparing the allelic composition between Mali and Nigeria, significant difference was found, suggesting high population differentiation may occur in the 2 endemic countries.
Applying microsatellite markers and different developmental stages for genotyping
S. haematobium, significant genetic diversity was determined in the populations in Africa where the disease prevalence varies among countries. By using
S. haematobium individual miracidium hatched with the eggs from patients in Mali, it was found that there was only limited evidence of population subdivision between individuals or sampling locations [
4]. Based on the microsatellite analysis on individual miracidium from patient’s egg samples, high levels of genetic diversity were detected in S. mansoni and
S. haematobium populations at the country level, but not at regional level, across 6 sub-Saharan African countries [
20]. Most recently, Glenn et al. [
23] developed highly variable DNA markers for individual adult worms of
S. haematobium from laboratory animals, and detected significant variance in genetic diversity and differentiation among populations of
S. haematobium in Africa. Our study demonstrated significant difference of genetic variation of
S. haematobium between the populations from Mali and Nigeria, but no evidence of population variance within Nigeria. The results support the previous findings that genetic variation exists in the
S. haematobium populations in Africa at country level, and it may be related to the varied prevalence of the disease in different countries.
We used pooled DNA of eggs from each patient, for the first time, to genotype natural
S. haematobium. An approach using pooled DNA of individual cloned adult worms of a laboratory strain was proposed for studying population genetics of
S. mansoni, presenting a good correlation between the pooled values and the true allele frequencies [
13]. Although this method can be used for larger scale of genotyping, it should be recognized that it has potential limitations. The pooled data could not be assessed by using Hardy-Weinberg probability and other parameters for population differentiation, and the estimation accuracy of genotype frequency is depending on how the pool is made. If a pool contains all parasites from each individual, sampling bias could be minimized. The number of eggs for DNA pool, samples, and the microsatellite loci was limited in our study, thus limit the power to estimate the population genetic diversity, and to reduce sampling bias. As commonly acknowledged, a best way to evaluate intra- and inter-population genetic structures and variations at the individual host level is to use DNA derived from single genotypes. Although there are shortcomings, the parasite egg stage, we used, could be a simpler and lower labour-consuming way to study genetic variation of
Schistosoma from the definitive host. However, when using pooled egg DNA to assess the allele frequency of parasites, the DNA pool should be prepared using all eggs or a larger number of eggs from each individual to reduce sampling bias. To evaluate population differentiation, individual eggs and more microsatellite loci should be applied for further study.
Notes
-
We have no conflict of interest related to this study.
We thank Dr. Lester Chitsulo from WHO (Switzerland), Prof. Xiaonong Zhou (National Institute of Parasitic Diseases, China), for the discussion and critical comments. This research was funded by the National Science & Technology Priority Program, China (no. 2012ZX10004-220).
Fig. 1.Location of S. haematobium samples collected.
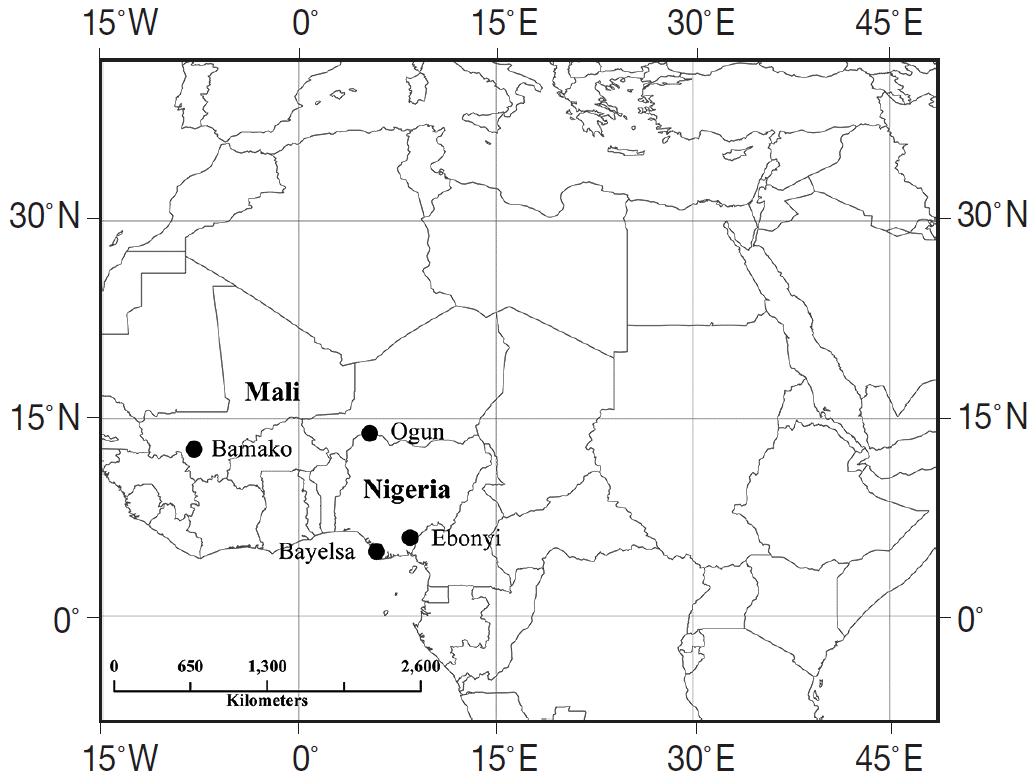
Fig. 2.Comparison of allelic composition among 3 locations in Nigeria.
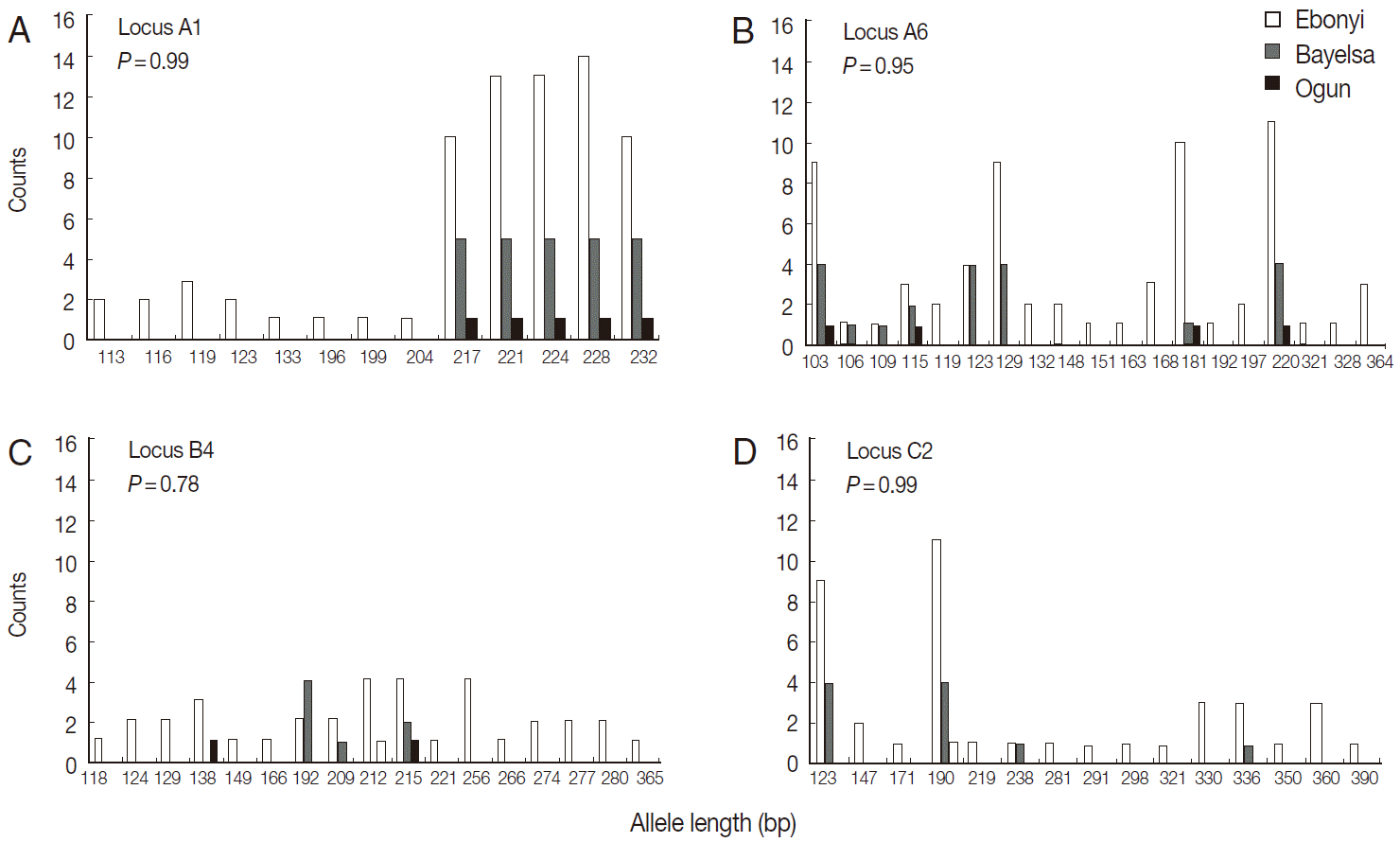
Fig. 3.Comparison of allelic composition between Mali and Nigerian populations.

Table 1.Allele counts of S. haemabobium from 4 locations in Africa
Table 1.
|
Locus |
|
Mali |
Nigeria
|
|
|
Locus |
|
Mali |
Nigeria
|
|
Ebonyi |
Bayelsa |
Ogun |
Subtotal |
|
|
|
Ebonyi |
Bayelsa |
Ogun |
Subtotal |
|
No. patients |
20 |
16 |
5 |
1 |
22 |
|
|
|
No. patients |
27 |
14 |
5 |
1 |
20 |
|
A1 |
110 |
2 |
0 |
0 |
0 |
0 |
|
|
B4 |
118 |
6 |
1 |
0 |
0 |
1 |
|
113 |
2 |
2 |
0 |
0 |
2 |
|
|
|
124 |
0 |
2 |
0 |
0 |
2 |
|
116 |
1 |
2 |
0 |
0 |
2 |
|
|
|
129 |
2 |
2 |
0 |
0 |
2 |
|
119 |
0 |
3 |
0 |
0 |
3 |
|
|
|
138 |
1 |
3 |
0 |
1 |
4 |
|
123 |
0 |
2 |
0 |
0 |
2 |
|
|
|
146 |
3 |
0 |
0 |
0 |
0 |
|
133 |
0 |
1 |
0 |
0 |
1 |
|
|
|
149 |
0 |
1 |
0 |
0 |
1 |
|
196 |
0 |
1 |
0 |
0 |
1 |
|
|
|
166 |
1 |
1 |
0 |
0 |
1 |
|
199 |
0 |
1 |
0 |
0 |
1 |
|
|
|
192 |
3 |
2 |
4 |
0 |
6 |
|
204 |
0 |
1 |
0 |
0 |
1 |
|
|
|
209 |
22 |
2 |
1 |
0 |
3 |
|
217 |
18 |
10 |
5 |
1 |
16 |
|
|
|
212 |
22 |
4 |
0 |
1 |
5 |
|
221 |
19 |
13 |
5 |
1 |
19 |
|
|
|
215 |
24 |
4 |
2 |
1 |
7 |
|
224 |
20 |
13 |
5 |
1 |
19 |
|
|
|
221 |
0 |
1 |
0 |
0 |
1 |
|
228 |
20 |
14 |
5 |
1 |
20 |
|
|
|
249 |
1 |
0 |
0 |
0 |
0 |
|
232 |
19 |
10 |
5 |
1 |
16 |
|
|
|
256 |
0 |
4 |
0 |
0 |
4 |
|
No. patients |
27 |
16 |
4 |
1 |
21 |
|
|
|
266 |
0 |
1 |
0 |
0 |
1 |
|
A6 |
103 |
21 |
9 |
4 |
1 |
14 |
|
|
|
274 |
18 |
2 |
0 |
0 |
2 |
|
106 |
9 |
1 |
1 |
0 |
2 |
|
|
|
277 |
18 |
2 |
0 |
0 |
2 |
|
109 |
6 |
1 |
1 |
0 |
2 |
|
|
|
280 |
18 |
2 |
0 |
0 |
2 |
|
115 |
14 |
3 |
2 |
1 |
6 |
|
|
|
340 |
8 |
0 |
0 |
0 |
0 |
|
119 |
11 |
2 |
0 |
0 |
2 |
|
|
|
346 |
8 |
0 |
0 |
0 |
0 |
|
123 |
12 |
4 |
4 |
0 |
8 |
|
|
|
365 |
0 |
1 |
0 |
0 |
1 |
|
129 |
15 |
9 |
4 |
0 |
13 |
|
|
|
No. patients |
27 |
16 |
5 |
1 |
22 |
|
132 |
1 |
2 |
0 |
0 |
2 |
|
|
C2 |
107 |
2 |
0 |
0 |
0 |
0 |
|
135 |
1 |
0 |
0 |
0 |
0 |
|
|
|
111 |
1 |
0 |
0 |
0 |
0 |
|
145 |
1 |
0 |
0 |
0 |
0 |
|
|
|
117 |
7 |
0 |
0 |
0 |
0 |
|
148 |
3 |
2 |
0 |
0 |
2 |
|
|
|
123 |
18 |
9 |
4 |
0 |
13 |
|
151 |
3 |
1 |
0 |
0 |
1 |
|
|
|
132 |
18 |
0 |
0 |
0 |
0 |
|
158 |
3 |
0 |
0 |
0 |
0 |
|
|
|
147 |
0 |
2 |
0 |
0 |
2 |
|
163 |
1 |
1 |
0 |
0 |
1 |
|
|
|
157 |
3 |
0 |
0 |
0 |
0 |
|
168 |
0 |
3 |
0 |
0 |
3 |
|
|
|
171 |
1 |
1 |
0 |
0 |
1 |
|
181 |
8 |
10 |
1 |
1 |
12 |
|
|
|
178 |
1 |
0 |
0 |
0 |
0 |
|
192 |
6 |
1 |
0 |
0 |
1 |
|
|
|
190 |
14 |
11 |
4 |
1 |
16 |
|
197 |
0 |
2 |
0 |
0 |
2 |
|
|
|
199 |
16 |
0 |
0 |
0 |
0 |
|
201 |
1 |
0 |
0 |
0 |
0 |
|
|
|
210 |
4 |
0 |
0 |
0 |
0 |
|
220 |
19 |
11 |
4 |
1 |
16 |
|
|
|
219 |
2 |
1 |
0 |
0 |
1 |
|
223 |
2 |
0 |
0 |
0 |
0 |
|
|
|
238 |
9 |
1 |
1 |
0 |
2 |
|
226 |
4 |
0 |
0 |
0 |
0 |
|
|
|
267 |
5 |
0 |
0 |
0 |
0 |
|
229 |
1 |
0 |
0 |
0 |
0 |
|
|
|
281 |
0 |
1 |
0 |
0 |
1 |
|
238 |
5 |
0 |
0 |
0 |
0 |
|
|
|
291 |
2 |
1 |
0 |
0 |
1 |
|
252 |
1 |
0 |
0 |
0 |
0 |
|
|
|
298 |
1 |
1 |
0 |
0 |
1 |
|
287 |
4 |
0 |
0 |
0 |
0 |
|
|
|
316 |
5 |
0 |
0 |
0 |
0 |
|
321 |
0 |
1 |
0 |
0 |
1 |
|
|
|
321 |
0 |
1 |
0 |
0 |
1 |
|
328 |
0 |
1 |
0 |
0 |
1 |
|
|
|
330 |
0 |
3 |
0 |
0 |
3 |
|
334 |
1 |
0 |
0 |
0 |
0 |
|
|
|
336 |
4 |
3 |
1 |
0 |
4 |
|
340 |
1 |
0 |
0 |
0 |
0 |
|
|
|
350 |
0 |
1 |
0 |
0 |
1 |
|
364 |
0 |
3 |
0 |
0 |
3 |
|
|
|
360 |
2 |
3 |
0 |
0 |
3 |
|
|
|
|
|
|
|
|
|
|
390 |
0 |
1 |
0 |
0 |
1 |
Table 2.Characteristics of microsatellite assay for S. haematobium from 4 locations in Africa
Table 2.
|
Allele character |
A1
|
A6
|
B4
|
C2
|
|
Mali |
Nigeria |
Mali |
Nigeria |
Mali |
Nigeria |
Mali |
Nigeria |
|
No. patients tested |
20 |
22 |
27 |
21 |
27 |
20 |
27 |
22 |
|
Average no. alleles |
5.1 |
4.7 |
5.8 |
4.3 |
5.9 |
2.3 |
4.3 |
2.3 |
|
Range of allele number |
2-7 |
1-8 |
2-10 |
1 -8 |
1-10 |
1 -8 |
1-9 |
1 -5 |
|
Range of allele size |
110-232 |
113-232 |
103-340 |
103-364 |
118-346 |
118-365 |
107-360 |
123-360 |
References
- 1. Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis 2006;6:411-425.
- 2. van der Werf MJ, de Vlas SJ, Brooker S, Looman CWN, Nagelkerke NJD, Habbema JDF, Engels D. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop 2003;86:125-139.
- 3. Brouwer KC, Ndhlovu PD, Wagatsuma Y, Munatsi A, Shiff CJ. Urinary tract pathology attributed to Schistosoma haematobium: does parasite genetics play a role? Am J Trop Med Hyg 2003;68:456-462.
- 4. Gower CM, Gabrielli AF, Sacko M, Dembele R, Golan R, Emery AM, Rollinson D, Webster JP. Population genetics of Schistosoma haematobium: development of novel microsatellite markers and their application to schistosomiasis control in Mali. Parasitology 2011;138:978-994.
- 5. Wright CA, Ross GC. Enzyme analysis of Schistosoma haematobium. Bull WHO 1983;61:307.
- 6. Littlewood DTJ, Lockyer AE, Webster BL, Johnston DA, Le TH. The complete mitochondrial genomes of Schistosoma haematobium and Schistosoma spindale and the evolutionary history of mitochondrial genome changes among parasitic flatworms. Mol Phylogen Evol 2006;39:452-467.
- 7. Ellegren H. Microsatellites: simple sequences with complex evolution. Nature Rev Genetics 2004;5:435-445.
- 8. Golan R, Gower CM, Emery AM, Rollinson D, Webster JP. Isolation and characterization of the first polymorphic microsatellite markers for Schistosoma haematobium and their application in multiplex reactions of larval stages. Mol Ecol Resour 2008;8:647-649.
- 9. Sorensen RE, Rodrigues NB, Oliveira G, Romanha AJ, Minchella DJ. Genetic filtering and optimal sampling of Schistosoma mansoni populations. Parasitology 2006;133:443-451.
- 10. Curtis J, Sorensen RE, Minchella DJ. Schistosome genetic diversity: the implications of population structure as detected with microsatellite markers. Parasitology 2002;125:S51-S59.
- 11. Pacek P, Sajantila A, Syvänen AC. Determination of allele frequencies at loci with length polymorphism by quantitative analysis of DNA amplified from pooled samples. Genom Res (PCR Meth Appl) 1993;2:313-317.
- 12. Shaw SH, Carrasquillo MM, Kashuk C, Puffenberger EG, Chakravarti A. Allele frequency distributions in pooled DNA samples: applications to mapping complex disease genes. Genome Res 1998;8:111-123.
- 13. Silva L, Liu S, Blanton RE. Microsatellite analysis of pooled Schistosoma mansoni DNA: an approach for studies of parasite populations. Parasitology 2006;132:331-338.
- 14. Reinstrup L, Jorgensen A, Vennervald BJ, Kristensen TK. DNA extraction from dried Schistosoma haematobium eggs isolated on nylon filters. Trans R Soc Trop Med Hyg 2012;106:270-272.
- 15. R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria. 2009.
- 16. Sokal RR, Rohlf FJ. Biometry. 3rd ed. San Francisco, USA. Freeman and Co; 1995.
- 17. Sham PC, Curtis D. Monte-Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet 1995;59:97-105.
- 18. Norton AJ, Gower CM, Lamberton PHL, Webster BL, Lwambo NJS, Blair L, Fenwick A, Webster JP. Genetic consequences of mass human chemotherapy for Schistosoma mansoni: population structure pre- and post-praziquantel treatment in Tanzania. Am J Trop Med Hyg 2010;83:951-957.
- 19. Webster BL, Emery AM, Webster JP, Gouvras A, Garba A, Diaw O, Seye MM, Tchuente LA, Simoonga C, Mwanga J, Lange CN, Kariuki C, Mohammed KA, Stothard JR, Rollinson D. Genetic diversity within Schistosoma haematobium: DNA barcoding reveals two distinct groups. PLoS Negl Trop Dis 2012;6:e1882.
- 20. Gower CM, Gouvras AN, Lamberton PHL, Deol A, Shrivastava J, Mutombo PN, Mbuh JV, Norton AJ, Webster BL, Stothard JR, Garba A, Lamine M, Kariuki C, Lang CN, Mkoji GM, Kabatereine NB, Gabrielli AF, Rudge JW, Fenwick A, Sacko M, Dembele R, Lwambo NJS, Tchuem LA, Rollinson D, Webster JP. Population genetic structure of Schistosoma mansoni and Schistosoma haematobium from across six sub-Saharan African countries: implications for epidemiology, evolution and control. Acta Trop 2013;128:261-274.
- 21. Rollinson D, Knopp S, Levitz S, Stothard JR, Tchuente L-AT, Garba A, Mohammed KA, Schur N, Person B, Colley DG, Utzinger J. Time to set the agenda for schistosomiasis elimination. Acta Trop 2013;128:423-440.
- 22. Whitehorn PR, Tinsley MC, Brown MJF, Darvill B, Goulson D. Genetic diversity, parasite prevalence and immunity in wild bumblebees. Proc Roy Soc B-Biol Sci 2011;278:1195-1202.
- 23. Glenn T, Lance S, McKee A, Webster BL, Emery E, Zerlotini A, Oliveira G, Rollinson D, Faircloth B. Significant variance in genetic diversity among populations of Schistosoma haematobium detected using microsatellite DNA loci from a genome-wide database. Parasit Vectors 2013;6:300.


