Abstract
Toxoplasma gondii is an opportunistic protozoan parasite that can infect almost all warm-blooded animals including humans with a worldwide distribution. Micronemes play an important role in invasion process of T. gondii, associated with the attachment, motility, and host cell recognition. In this research, sequence diversity in microneme protein 6 (MIC6) gene among 16 T. gondii isolates from different hosts and geographical regions and 1 reference strain was examined. The results showed that the sequence of all the examined T. gondii strains was 1,050 bp in length, and their A + T content was between 45.7% and 46.1%. Sequence analysis presented 33 nucleotide mutation positions (0-1.1%), resulting in 23 amino acid substitutions (0-2.3%) aligned with T. gondii RH strain. Moreover, T. gondii strains representing the 3 classical genotypes (Type I, II, and III) were separated into different clusters based on the locus of MIC6 using phylogenetic analyses by Bayesian inference (BI), maximum parsimony (MP), and maximum likelihood (ML), but T. gondii strains belonging to ToxoDB #9 were separated into different clusters. Our results suggested that MIC6 gene is not a suitable marker for T. gondii population genetic studies.
-
Key words: Toxoplasma gondii, toxoplamosis, MIC6, sequence diversity
Toxoplasma gondii is an apicomplexan parasite capable of infecting almost all warm-blooded animals and humans [
1,
2].
T. gondii infection could cause diverse diseases in immunocompromised patients and even infant birth defects in pregnant mothers [
3,
4].
T. gondii can also lead to abortion and fetal abnormality in livestock, resulting in serious economic losses to the farming industry [
5].
Microneme proteins (MICs) of
T. gondii play an important role in
T. gondii survival and invasion and thus affect host cell signaling [
6,
7]. During the invasion process, MICs participate in binding of
T. gondii to the host cell surface, as well as the formation of the bridge with the parasite actinomyosin system [
8,
9]. MIC6 is a member of the MIC4-MIC1-MIC6 complex, which mediates host cell recognition and attachment by the parasite [
10], but also it is a key factor of the parasite virulence [
11,
12]. Recent studies have demonstrated that different clonal types of
T. gondii isolates with diverse geographical distribution could cause different toxoplasmosis in animals and humans [
13,
14]. In order to uncover the details of
T. gondii genetic diversity, sequence variation in MIC6 gene among 16
T. gondii strains from different hosts and geographical locations and 1 reference isolate was examined in this research.
Sixteen
T. gondii strains belong to different genotypes from different hosts and geographical regions were used in this study (
Table 1) [
15-
17], and the MIC6 gene sequence of TgME49 strain (ToxoDB: TGME49_218520) was included for sequence analysis. According to MIC6 gene sequence of 3ME49 strain provided by ToxoDB database (
http://toxodb.org/toxo/), a pair of specific primers (forward primer, 5'-ATGAGGCTCTTCCGGTGCT-3'; reverse primer, 5'-TTAATCCCATGTTTTGCTATCC-3') was used to amplify MIC gene from individual isolate. The amplification reaction was carried out using Ex Taq polymerase (TaKaRa, Kyoto, Japan) according to the manufacturer’s recommendations. Amplification was performed in a thermocycler (Biometra, Gottingen, Germany) using the following protocol: denaturation at 94.0°C for 4 min followed by 35 cycles composing of 94.0˚C for 30 sec, 39.3˚C for 30 sec and 72.0˚C for 1 min, and a final extension step at 72.0˚C for 5 min. The confirmation of PCR amplifications by agarose gel electrophoresis was carried out as previously described [
18]. All the MIC6 PCR products were purified (Promega, Madison, Wisconsin, USA), and ligated with pMD18-T vector (TaKaRa) followed by transformed into JM109 competent cells (Promega) according to the manufacturer’s recommendations, the positive colonies were identified by PCR, and then sequenced as previously described [
18].
All the obtained MIC6 sequences were aligned using Clustal X 2.11 [
19], and evolutionary analysis was processed by MEGA 5.2 [
20]. The intra-specific sequence variation was evaluated by percent of the different bases. Phylogenetic reconstructions of the examined
T. gondii strains based on MIC6 gene sequence were performed by 3 inference methods, namely Bayesian inference (BI), maximum parsimony (MP), and maximum likelihood (ML) methods, using
Neospora caninum (
http://toxodb.org/toxo/: NCLIV_061760) as an out-group. BI analysis was carried out with 4 independent Markov chains run for 200,000 metropolis-coupled MCMC generations, sampling a tree every 100 generations in MrBayes 3.1.1 [
21]. Both MP and ML analyses were carried out using PAUP
* 4.0b10 [
22]. Bootstrap probability (BP) and random addition searches were performed as previously described [
23].
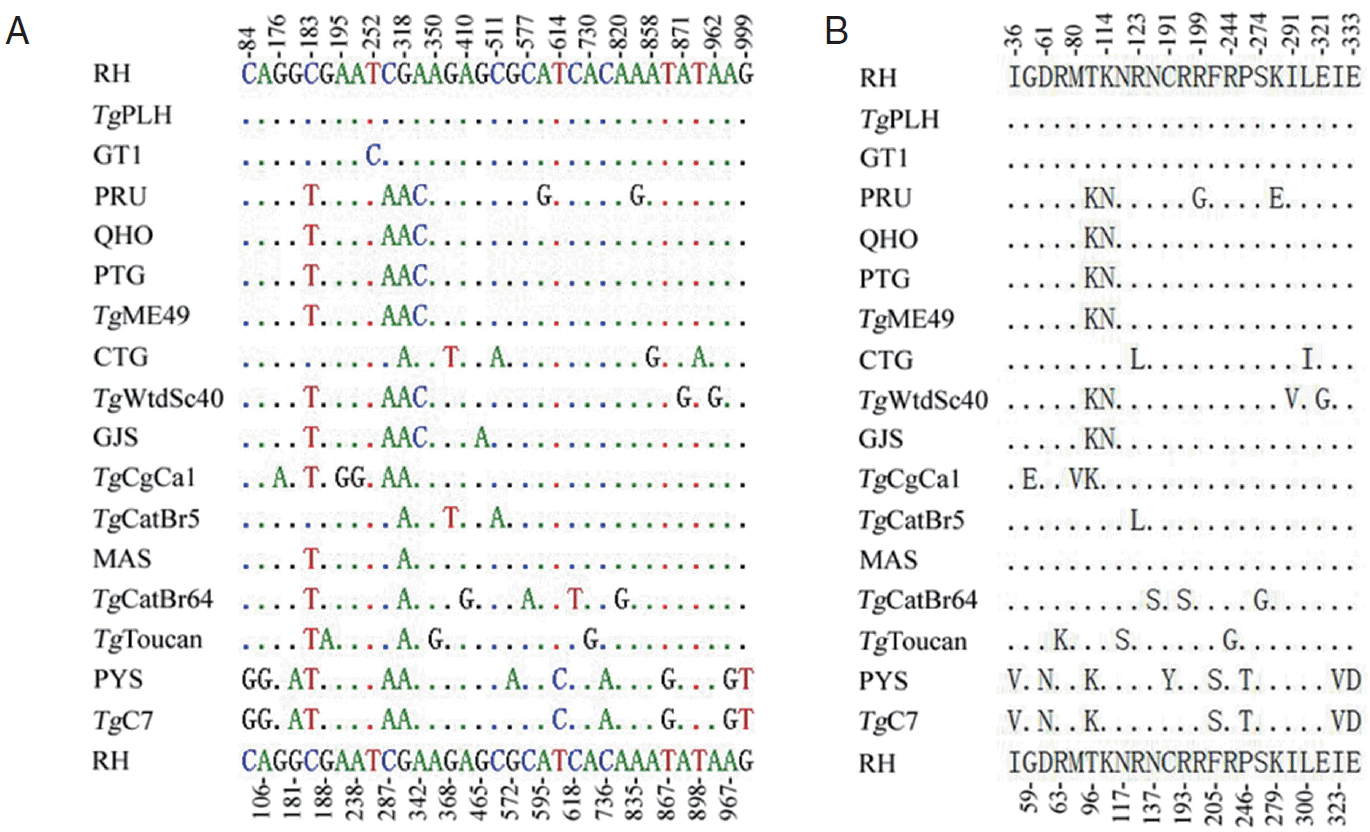
PCR amplification of MIC6 gene from individual
T. gondii isolates produced a single band of approximately 1,000 bp in length on agarose gel (not shown). Positive MIC6 transformants of expected length selected by PCR were sequenced from both directions. The length of the obtained MIC6 gene sequences for all the examined strains was 1,050 bp, and their A+T content ranged from 45.7% to 46.1%. Comparison of the obtained 16 MIC6 sequences plus that of the
TgME49 strain (ToxoDB: TGME49_218520) revealed nucleotide polymorphisms at 33 nucleotide positions (0-1.1%) (
Fig. 1A), which was similar to that in ROP38 [
24] but lower than that in GRA5 [
23] and GRA6 [
25]. There were 23 amino acid substitutions (0-2.3%) (
Fig. 1B) due to the nucleotide mutation (33 mutations) compared with
T. gondii RH isolate. 22 transitions (A↔G and C↔T) and 18 transversions (A↔C, A↔T, G↔T and G↔C) were identified among the examined
T. gondii strains, and the distance of evolutionary divergence was 0-8.7%, suggesting that the variation rate was low in MIC6 gene among the examined
T. gondii isolates. Thus, our data has supported previous studies that MIC6 is a potential vaccine candidate against
T. gondii RH [
11] and PRU infections [
12].
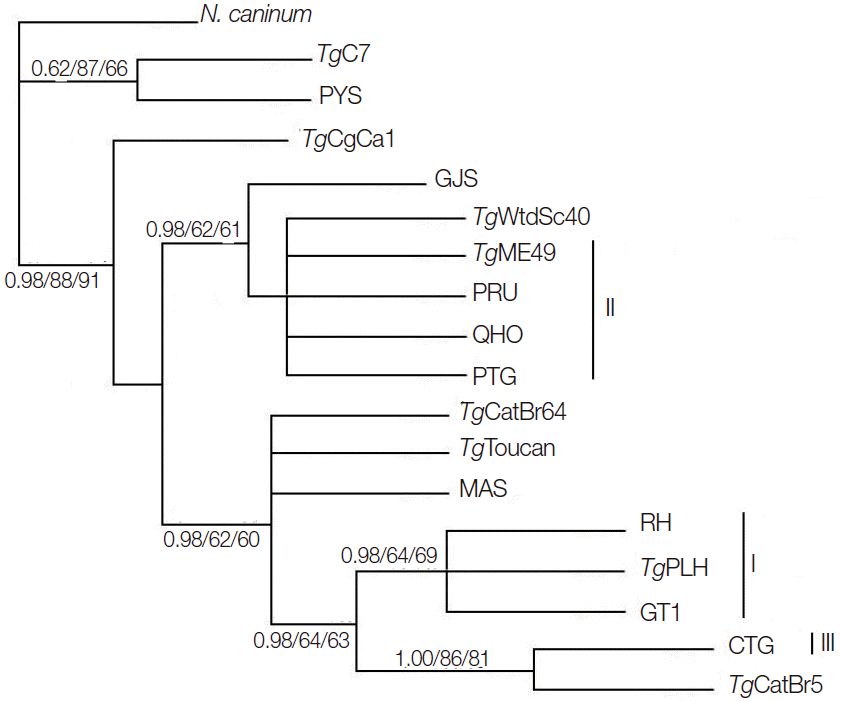
Phylogenetic reconstruction of the examined
T. gondii strains using BI, MP, and ML methods is shown in
Fig. 2.
T. gondii strains representing the 3 classical genotypes (Type I, II, and III) were separated into different clusters, but
T. gondii strains belonging to ToxoDB #9 were separated into different clusters. This result is similar to that using GRA5 [
23] and eIF4A [
18] as genetic markers, suggesting that
MIC6 gene may not represent a suitable marker for
T. gondii population genetic studies.
Notes
-
The authors declare that they have no conflict of interest.
The project support was provided, in part, by the National Science Foundation of China (grant no. 31228022), and The Science Fund for Creative Research Groups of Gansu Province (grant no. 1210RJIA006).
Fig. 1.Multiple alignment analyses of nucleotides (A) or amino acid sequences (B) of Toxoplasma gondii MIC6 gene. Point (.) indicates identical nucleotide or amino acid compared with that of T. gondii RH strain (upper and bottom lines), and the number indicates the variable sequence positions for nucleotide (A) or amino acid (B).
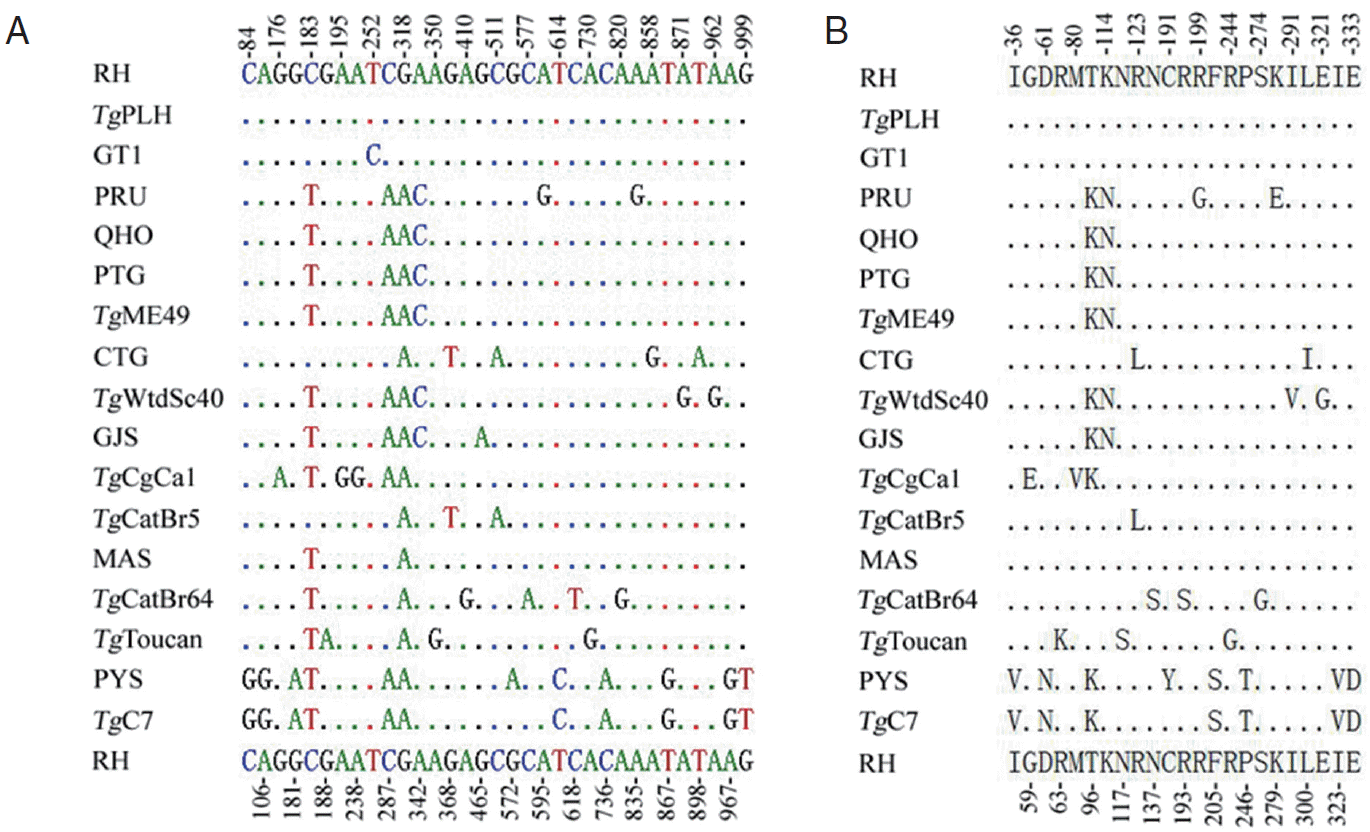
Fig. 2.Phylogenetic analysis of 17 Toxoplasma gondii strains (including TgME49) based on MIC6 gene sequences using Bayesian inference (BI), maximum parsimony (MP), and maximum likelihood (ML) methods, with N. caninum as the out-group. Numbers nearby the branches indicate bootstrap values from different analysis in the order of BI, MP, and ML, and clusters of 3 classical genotypes were denoted by I, II, and III, respectively.
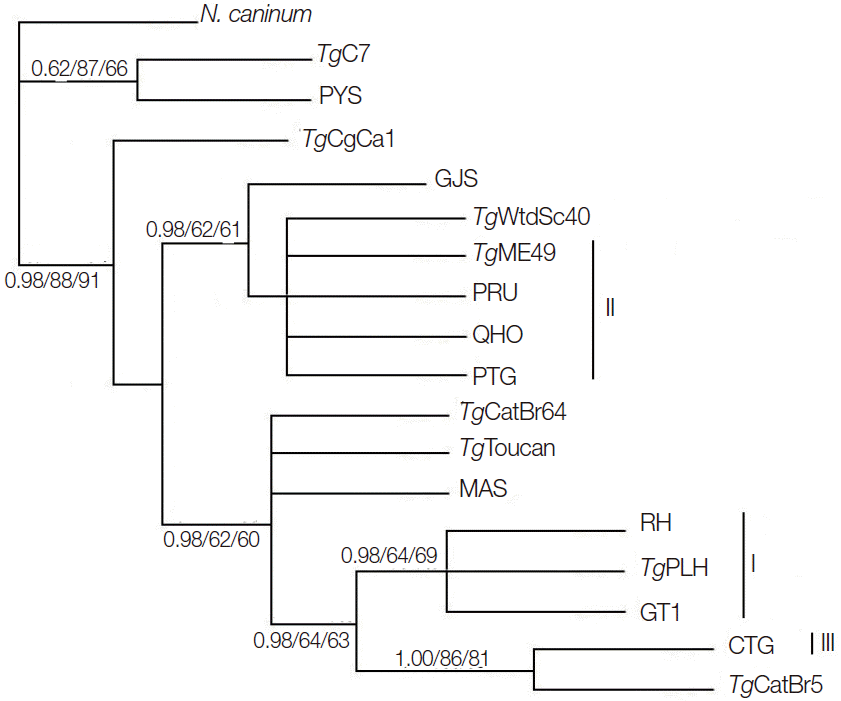
Table 1.Details of Toxoplasma gondii strains used in this study
Table 1.
|
No. |
Isolate |
Host |
Geographical Location |
Genotypea
|
|
1 |
RH |
Human |
France |
Reference, Type I, ToxoDB #10 |
|
2 |
TgPLH |
Pig |
Henan, China |
Type I, ToxoDB #10 |
|
3 |
GT1 |
Goat |
United States |
Reference, Type I, ToxoDB#10 |
|
4 |
MAS |
Human |
France |
Reference, ToxoDB#17 |
|
5 |
TgCgCa1 |
Cougar |
Canada |
Reference, ToxoDB#66 |
|
6 |
TgCatBr64 |
Cat |
Brazil |
Reference, ToxoDB#111 |
|
7 |
TgCatBr5 |
Cat |
Brazil |
Reference, ToxoDB#111 |
|
8 |
PRU |
Human |
France |
Type II, ToxoDB #1 |
|
9 |
QHO |
Sheep |
Qinghai, China |
Type II, ToxoDB #1 |
|
10 |
PTG |
Sheep |
United States |
Reference, Type II, ToxoDB#1 |
|
11 |
TgC7 |
Cat |
Guangzhou, China |
ToxoDB #9 |
|
12 |
PYS |
Pig |
Panyu, China |
ToxoDB #9 |
|
13 |
GJS |
Pig |
Jingyuan, Gansu, China |
ToxoDB #9 |
|
14 |
CTG |
Cat |
United States |
Reference, Type III, ToxoDB#2 |
|
15 |
TgWtdSc40 |
Deer |
USA |
Type 12, ToxoDB#5 |
|
16 |
TgToucan |
Toucan |
Costa Rica |
Reference, ToxoDB#52 |
References
- 1. Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet 2004;363:1965-1976.
- 2. Schlüter D, Däubener W, Schares G, Groß U, Pleyer U, Lüder C. Animals are key to human toxoplasmosis. Int J Med Microbiol 2014;304:917-929.
- 3. Kim K, Weiss LM. Toxoplasma: the next 100 years. Microbes Infect 2008;10:978-984.
- 4. Amin A, Mazloomzadeh S, Haniloo A, Mohammadian F, Fazaeli A. Evaluation of anti-Toxoplasma IgG, IgM, and IgA in mothers with spontaneous abortion in Zanjan, Northwest Iran. Korean J Parasitol 2012;50:371-374.
- 5. Dubey JP, Hill DE, Jones JL, Hightower AW, Kirkland E, Roberts JM, Marcet PL, Lehmann T, Vianna MC, Miska K, Streekumar C, Kwok OC, Shen SK, Gamble HR. Prevalence of viable Toxoplasma gondii in beef, chicken, and pork from retail meat stores in the United States: risk assessment to consumers. J Parasitol 2005;91:1082-1093.
- 6. Muniz-Feliciano L, Van Grol J, Portillo JA, Liew L, Liu B, Carlin CR, Carruthers VB, Matthews S, Subauste CS. Toxoplasma gondii-induced activation of EGFR prevents autophagy protein-mediated killing of the parasite. PLoS Pathog 2013;9:e1003809.
- 7. Soldati D, Meissner M. Toxoplasma as a novel system for motility. Curr Opin Cell Biol 2004;16:32-40.
- 8. Carruthers VB, Giddings OK, Sibley LD. Secretion of micronemal proteins is associated with Toxoplasma invasion of host cells. Cell Microbiol 1999;1:225-235.
- 9. Jewett TJ, Sibley LD. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol Cell 2003;11:885-894.
- 10. Zheng B, He A, Gan M, Li Z, He H, Zhan X. MIC6 associates with aldolase in host cell invasion by Toxoplasma gondii. Parasitol Res 2009;105:441-445.
- 11. Peng GH, Yuan ZG, Zhou DH, He XH, Liu MM, Yan C, Yin CC, He Y, Lin RQ, Zhu XQ. Toxoplasma gondii microneme protein 6 (MIC6) is a potential vaccine candidate against toxoplasmosis in mice. Vaccine 2009;27:6570-6574.
- 12. Yan HK, Yuan ZG, Song HQ, Petersen E, Zhou Y, Ren D, Zhou DH, Li HX, Lin RQ, Yang GL, Zhu XQ. Vaccination with a DNA vaccine coding for perforin-like protein 1 and MIC6 induces significant protective immunity against Toxoplasma gondii. Clin Vaccine Immunol 2012;19:684-689.
- 13. Sibley LD, Ajioka JW. Population structure of Toxoplasma gondii: clonal expansion driven by infrequent recombination and selective sweeps. Ann Rev Microbiol 2008;62:329-351.
- 14. Robert-Gangneux F, Dardé ML. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev 2012;25:264-296.
- 15. Zhou P, Zhang H, Lin RQ, Zhang DL, Song HQ, Su C, Zhu XQ. Genetic characterization of Toxoplasma gondii isolates from China. Parasitol Int 2009;58:193-195.
- 16. Zhou P, Nie H, Zhang LX, Wang HY, Yin CC, Su C, Zhu XQ, Zhao JL. Genetic characterization of Toxoplasma gondii isolates from pigs in China. J Parasitol 2010;96:1027-1029.
- 17. Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology 2010;137:1-11.
- 18. Chen J, Fang SF, Zhou DH, Li ZY, Liu GH, Zhu XQ. Sequence variation in the Toxoplasma gondii eIF4A gene among strains from different hosts and geographical locations. Genet Mol Res 2014;13:3356-3361.
- 19. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;25:4876-4882.
- 20. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28:2731-2739.
- 21. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003;19:1572-1574.
- 22. Swofford DL. Paup*: phylogenetic analysis using parsimony, version 4.0b10. Sunderland, Massachusetts, USA. Sinauer Associates; 2002.
- 23. Chen J, Li ZY, Zhou DH, Liu GH, Zhu XQ. Genetic diversity among Toxoplasma gondii strains from different hosts and geographical regions revealed by sequence analysis of GRA5 gene. Parasit Vectors 2012;5:279.
- 24. Xu Y, Zhang NZ, Chen J, Liu GH, Xu QM, Zhou DH, Zhu XQ. Toxoplasma gondii rhoptry protein 38 gene: sequence variation among isolates from different hosts and geographical locations. Genet Mol Res 2014;13:4839-4844.
- 25. Fazaeli A, Carter PE, Darde ML, Pennington TH. Molecular typing of Toxoplasma gondii strains by GRA6 gene sequence analysis. Int J Parasitol 2000;30:637-642.
Citations
Citations to this article as recorded by