Abstract
Toxoplasma gondii is a eukaryotic parasite of the phylum Apicomplexa, which infects all warm-blood animals, including humans. In the present study, we examined sequence variation in dense granule 20 (GRA20) genes among T. gondii isolates collected from different hosts and geographical regions worldwide. The complete GRA20 genes were amplified from 16 T. gondii isolates using PCR, sequence were analyzed, and phylogenetic reconstruction was analyzed by maximum parsimony (MP) and maximum likelihood (ML) methods. The results showed that the complete GRA20 gene sequence was 1,586 bp in length among all the isolates used in this study, and the sequence variations in nucleotides were 0-7.9% among all strains. However, removing the type III strains (CTG, VEG), the sequence variations became very low, only 0-0.7%. These results indicated that the GRA20 sequence in type III was more divergence. Phylogenetic analysis of GRA20 sequences using MP and ML methods can differentiate 2 major clonal lineage types (type I and type III) into their respective clusters, indicating the GRA20 gene may represent a novel genetic marker for intraspecific phylogenetic analyses of T. gondii.
-
Key words: Toxoplasma gondii, sequence variation, dense granule 20 (GRA20), phylogenetic analysis
Toxoplasma gondii, one of the most successful intracellular protozoan parasites, can infect the majority of vertebrate spices including humans with a worldwide distribution [
1-
3], and approximately one-third of the population has been exposed to
T. gondii. Normally, the infections are asymptomatic or subclinical. However, the
T. gondii infection can cause abortion and stillbirth in pregnant women, and encephalitis, chorioretinitis, and systemic infections in immunocompromised individuals [
2]. In animals,
T. gondii can also cause abortion in livestock, especially in sheep and goats, which can spawn a great number of economic losses in livestock [
3]. However, there was no effective vaccine and drugs that can help to control toxoplasmosis.
The strains of
T. gondii that predominate in Europe and North America, classified into types I, II, and III, differ in a wide range of phenotypes, including virulence, persistence, migratory capacity, and how they interface with the immune response [
4-
6], Thus, the information of genetic diversity of
T. gondii is useful for better understanding epidemiological patterns and pathogenicity, as well as exploring of new polymorphic virulence effectors.
GRA20, a novel dense granule protein, is secreted and targeted to parasitophorous vacuole membrane (PVM), which may participate in the manipulation of the host immunity [
7]. Previous studies have identified the existence of polymorphisms in dense granule proteins, such as GRA15, GRA5, and GRA6 [
8-
10], but the sequence variation about the
GRA20 gene among different
T. gondii isolates is still unknown. Therefore, the objective of this study was to examine sequence diversity of
GRA20 gene among
T. gondii strains from different hosts and geographical regions worldwide.
In this study, a total 16
T. gondii strains from different hosts and geographic locations were used for analysis (
Table 1). These
T. gondii isolates have been genotyped and genomic DNA (gDNA) was prepared as described previously [
11-
13].
To acquire amplicons of GRA20 genes concerning different T. gondii isolates, the primers GRA20-F (5´- ATGCATAGCCGGAACTGCGTC-3´) and GRA20-R (5´- TCACGCGGGCTTTCTACGG-3´) were designed based on T. gondii ME49 strain available in ToxoDB database (TGME49_200010). All the PCR products of GRA20 genes were purified by the DNA purification kit (GenStar, Beijing, China), ligated into pMD18-T vector (TaKaRa, Dalian, China), and then transformed into JM109 competent cells (Promega, Madison, Wisconsin, USA). Subsequently, the positive colonies were screened by PCR, and then sequenced by GenScript Co., Ltd. (Nanjing, China).
The acquired
GRA20 gene sequences were aligned by the Multiple Sequence Alignment Program, Clustal X 1.83 [
14], and sequence variation was determined among the examined
T. gondii strains. Phylogenetic reconstructions based on the complete sequences of
GRA20 gene among 13
T. gondii isolates and plus the corresponding sequences of strains TgCatBr9, VEG, and ME49 available in ToxoDB (
http://toxodb.org/toxo/) were carried out by 2 inference methods, maximum likelihood (ML) and maximum parsimony (MP) methods by Paup, with the sequence of
Hammondia hammondi (XM_008890671.1) as the out-group. Phylograms were drawn by the Tree View program version 1.65.
In the present study, the obtained entire genomic sequences of the GRA20 gene were 1,586 bp in length in all examined isolates. According to the analysis of all the 16 GRA20 complete sequences, there were 2 extrons and 1 intron in the
GRA20 gene (
Table 2). The A+T content ranged from 45.0% to 45.4% in the entire sequence. There were 124 nucleotide position variations with a distribution of 57 transitions (A↔G and C↔T), 61 transversions (C↔G, T↔G, A↔C, and A↔T) in CDS, and 2 transitions (A↔G and C↔T), and 1 transversion (C↔G, T↔G, A↔C, and A↔T) in the intron (
Table 2). However, when we analyzed the
GRA20 sequences without type III (CTG, VEG) strains, there were 12 nucleotide variations with a distribution of 10 transitions (A↔G and C↔T), 2 transversions (C↔G, T↔G, A↔C, and A↔T) in CDS, and 2 transitions (A↔G and C↔T) and 1 transversion (C↔G, T↔G, A↔C, and A↔T) in the intron. The alignment of
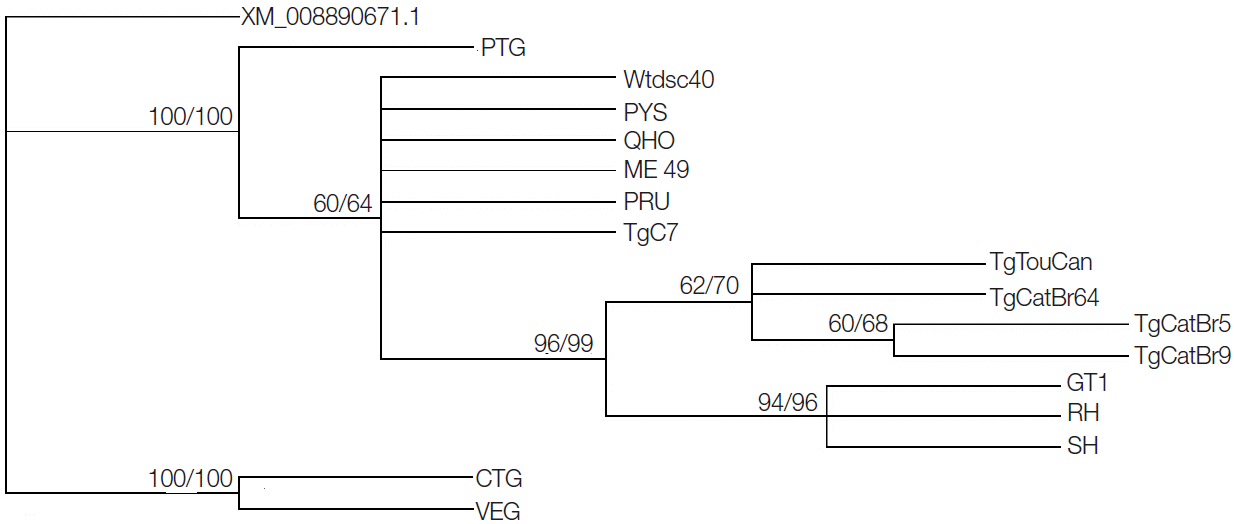
GRA20 gene sequences showed that sequence variation was 0-7.9% in all studied strains, while the sequence variation became 0-0.7% without the CTG and VEG strains. Phylogenetic reconstruction of all 16
T. gondii strains based on
GRA20 sequence data showed that the type I and type III of
T. gondii strains were clustered into respective clusters separately (
Fig. 1).
Recently, polymorphisms in the sequences of
GRA5, GRA6, GRA7, and
GRA15 genes have been reported [
8,
9,
15,
16]. Among them, polymorphic dense granule proteins were widely used in typing
T. gondii isolates, such as GRA6 [
10]. Furthermore, polymorphic dense granule protein may have different roles in regulating the inflammatory response. For example, GRA15 in type II activate more IL-12 than type I or type III strains [
8]. In this study, we found
GRA20 gene was very diverse in type III, indicating the functions may be different, too. Our results were consistent with that of some previous studies using other genetic markers, such as
GRA5, Rop17, and
HSP60 for genotyping [
9,
17,
18], but different to some previous studies, such as
Rop38 and
eIF4A [
19,
20].
In conclusion, the present study examined the sequences of the T. gondii GRA20 gene and revealed that it was more divergence in type III compared to other T. gondii strains, suggesting the functions of GRA20 in type III may be different from other strains. Phylogenetic analysis indicated that the GRA20 gene could distinguish the type I and type III strains, suggesting the GRA20 gene may be a novel genetic marker for studying genetic variation or the population genetic structures of T. gondii isolates.
Notes
-
The authors declare that they have no competing interests.
The project support was provided by National Natural Science Foundation of China (Grant No. 31228022) and the Science Fund for Creative Research Groups of Gansu Province (Grant No. 1210RJIA006).
Fig. 1.Phylogram of 16 Toxoplasma gondii isolates determined by analysis of the entire sequences of the GRA20 gene. The tree was reconstructed by maximum parsimony (MP) and maximum likelihood (ML) analyses. The numbers along branches indicate bootstrap values resulting from different analyses in the order: MP/ML.
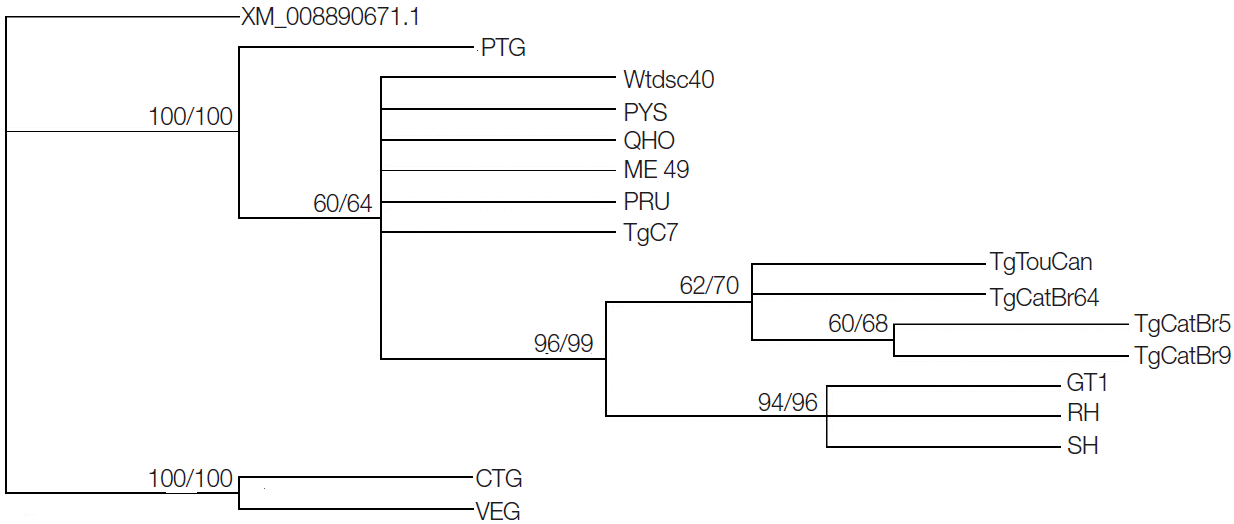
Table 1.Details of Toxoplasma gondii isolates used in this research
Table 1.
|
Isolate |
Host |
Geographical location |
Genotypea
|
|
RH |
Human |
France |
Reference, type I, ToxoDB #10 |
|
GT1 |
Goat |
United States |
Reference, type I, ToxoDB#10 |
|
SH |
Human |
Shanghai, China |
type I, ToxoDB #10 |
|
TgCatBr9 |
Cat |
Brazil |
ToxoDB#42 |
|
VEG |
Human |
United States |
Reference, ToxoDB#2 |
|
ME49 |
Sheep |
United States |
type II, ToxoDB#1 |
|
TgCatBr64 |
Cat |
Brazil |
Reference, ToxoDB#111 |
|
TgCatBr5 |
Cat |
Brazil |
Reference, ToxoDB#19 |
|
PRU |
Human |
France |
type II, ToxoDB #1 |
|
QHO |
Sheep |
Qinghai, China |
type II, ToxoDB #1 |
|
PTG |
Sheep |
United States |
Reference, type II, ToxoDB#1 |
|
TgC7 |
Cat |
Guangzhou, China |
ToxoDB #9 |
|
PYS |
Pig |
Panyu, China |
ToxoDB #9 |
|
CTG |
Cat |
United States |
Reference, type III, ToxoDB#2 |
|
TgWtdSc40 |
Deer |
USA |
type 12, ToxoDB#5 |
|
TgToucan |
Toucan |
Costa Rica |
Reference, ToxoDB#52 |
Table 2.Characteristics of Toxoplasma gondii GRA20 (TgGRA20) gene sequences
Table 2.
|
Item |
DNA
|
CDSa
|
First Extron
|
First Intron
|
Second Extron
|
|
ALLb
|
Except IIIc
|
ALL |
Except III |
ALL |
Except III |
ALL |
Except III |
ALL |
Except III |
|
Length (bp) |
1,586 |
1,586 |
1,242 |
1,242 |
140 |
140 |
344 |
344 |
1,102 |
1,102 |
|
T+A (%) |
44.96-45.40 |
45.02-45.40 |
42.83-43.32 |
42.91-43.32 |
45.71-46.43 |
45.71-46.43 |
52.62-52.91 |
52.62-52.91 |
42.47-42.92 |
42.56-42.92 |
|
Transition |
59 |
12 |
57 |
10 |
1 |
1 |
2 |
2 |
56 |
9 |
|
A↔G |
33 |
7 |
31 |
5 |
1 |
1 |
2 |
2 |
30 |
4 |
|
C↔T |
26 |
5 |
26 |
5 |
/ |
/ |
/ |
/ |
26 |
5 |
|
Transversion |
62 |
3 |
61 |
2 |
0 |
0 |
1 |
1 |
61 |
2 |
|
A↔T |
8 |
/ |
8 |
/ |
/ |
/ |
/ |
/ |
8 |
/ |
|
G↔C |
17 |
1 |
17 |
1 |
/ |
/ |
/ |
/ |
17 |
1 |
|
A↔C |
19 |
2 |
18 |
1 |
/ |
/ |
1 |
1 |
18 |
1 |
|
G↔T |
18 |
/ |
18 |
/ |
/ |
/ |
/ |
/ |
18 |
/ |
|
loss |
6 |
0 |
6 |
0 |
0 |
0 |
0 |
0 |
6 |
0 |
|
VNd
|
127 |
15 |
124 |
12 |
1 |
1 |
3 |
3 |
123 |
11 |
|
Re
|
0.95 |
4 |
0.93 |
5 |
∖ |
∖ |
2 |
2 |
0.92 |
4.5 |
|
Distance (%) |
0-7.9 |
0-0.7 |
0-10.1 |
0-0.6 |
0-0.7 |
0-0.7 |
0-0.9 |
0-0.9 |
0-11.4 |
0-0.6 |
References
- 1. Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet 2004;363:1965-1976.
- 2. Weiss LM, Dubey JP. Toxoplasmosis: a history of clinical observations. Int J Parasitol 2009;39:895-901.
- 3. Dubey JP. Toxoplasmosis of animals and humans. Boca Raton, Florida, USA. CRC Press; 2010.
- 4. Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis 1995;172:1561-1566.
- 5. Melo MB, Jensen KD, Saeij JP. Toxoplasma gondii effectors are master regulators of the inflammatory response. Trends Parasitol 2011;27:487-495.
- 6. Hunter CA, Sibley LD. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat Rev Microbiol 2012;10:766-778.
- 7. Hsiao CHC, Luisa Hiller N, Haldar K, Knoll LJ. A HT/PEXEL motif in Toxoplasma dense granule proteins is a signal for protein cleavage but not export into the host cell. Traffic 2013;14:519-531.
- 8. Rosowski EE, Lu D, Julien L, Rodda L, Gaiser RA, Jensen KD, Saeij JP. Strain-specific activation of the NF-kB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J Exp Med 2011;208:195-212.
- 9. Chen J, Li ZY, Zhou DH, Liu GH, Zhu XQ. Genetic diversity among Toxoplasma gondii strains from different hosts and geographical regions revealed by sequence analysis of GRA5 gene. Parasit Vectors 2012;5:279.
- 10. Fazaeli A, Carter PE, Pennington TH. Molecular typing of Toxoplasma gondii strains by GRA6 gene sequence analysis. Int J Parasitol 2000;30:637-642.
- 11. Zhou P, Zhang H, Lin RQ, Zhang DL, Song HQ, Su C, Zhu XQ. Genetic characterization of Toxoplasma gondii isolates from China. Parasitol Int 2009;58:193-195.
- 12. Zhou P, Nie H, Zhang LX, Wang HY, Yin CC, Su C, Zhu XQ, Zhao JL. Genetic characterization of Toxoplasma gondii isolates from pigs in China. J Parasitol 2010;96:1027-1029.
- 13. Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology 2010;137:1-11.
- 14. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: fexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;25:4876-4882.
- 15. Sousa S, Ajzenberg D, Marle M, Aubert D, Villena I, da Costa JC, Dardé ML. Selection of polymorphic peptides from GRA6 and GRA7 sequences of Toxoplasma gondii strains to be used in serotyping. Clin Vaccine Immunol 2009;16:1158. –1169.
- 16. Ma JS, Sasai M, Ohshima J, Lee Y, Bando H, Takeda K, Yamamoto M. Selective and strain-specifc NFAT4 activation by the Toxoplasma gondii polymorphic dense granule protein GRA6. J Exp Med 2014;211:2013-2032.
- 17. Zhang NZ, Xu Y, Huang SY, Zhou DH, Wang RA, Zhu XQ. Sequence variation in Toxoplasma gondii rop17 gene among strains from different hosts and geographical locations. Scientific World Journal 2014;2014:349325.
- 18. Lu J, Zhou DH, Chen J, Zhang NZ, Wang RA, Weng YB, Zhu XQ. Characterization of the Toxoplasma gondii hsp60 gene sequences from different hosts and geographical locations. Genet Mol Res 2014;13:6906-6911.
- 19. Xu Y, Zhang NZ, Chen J, Liu GH, Xu QM, Zhou DH, Zhu XQ. Toxoplasma gondii rhoptry protein 38 gene: sequence variation among isolates from different hosts and geographical locations. Genet Mol Res 2014;13:4839-4844.
- 20. Chen J, Fang SF, Zhou DH, Li ZY, Liu GH, Zhu XQ. Sequence variation in the Toxoplasma gondii eIF4A gene among strains from different hosts and geographical locations. Genet Mol Res 2014;13:3356-3361.
Citations
Citations to this article as recorded by

- Effects of latent infection of Toxoplasma gondii strains with different genotypes on mouse behavior and brain transcripts
Bei-Bei Zhou, Hong-Jie Dong, Hang Sun, Xiao-Man Xie, Huan-Huan Xie, Wen-Ju Zhu, Ya-Nan Li, Chao Xu, Jian-Ping Cao, Gui-Hua Zhao, Kun Yin
Parasites & Vectors.2025;[Epub] CrossRef - First Report on the Molecular Detection and Genetic Characterization of Toxoplasma gondii From Donkeys in Kenya
Fredrick O. Obonyo, Ndichu Maingi, Samuel M. Githigia, Kevin O. Ochwedo, Anne A. Owiti, Evans N. Nyaboga
Acta Parasitologica.2024; 69(3): 1480. CrossRef