Abstract
The taxonomy of Spirometra species has been controversial despite the medical and veterinary importance. Currently, only a few Spirometra species are considered valid species in the genus Spirometra. In the present study, the distribution of Spirometra species obtained from animals in Korea were identified by molecular analysis of the mitochondrial cytochrome c oxidase I (cox1) gene. A total of 28 Spirometra species specimens were analyzed. These were all collected between 1973 and 2008 in the Republic of Korea. Mitochondrial cox1 sequences were examined for a total of 28 specimens comprising 14 S. decipiens and 14 S. ranarum. The difference in partial cox1 sequences (316 bp) between S. erinaceieuropaei (KJ599680) and S. ranarum (this study) was 9.3%, while that between S. decipiens (KJ599679) and S. ranarum (this study) was 2.2%. Genetic analyses identified 2 Spirometra species in animals such as cat, leopard cat, dog, duck and snake in Korea as S. decipiens and S. ranarum. S. decipiens and S. ranarum were present in Gyeongnam Province (P), Jeonnam P, Gangwon P, Chungbuk P, and Seoul. S. decipiens was found in tadpoles, snakes, ducks, cats, leopard cats and dogs, while S. ranarum was found in cats and dogs. The ratio of S. decipiens:S. ranarum calculated from the molecular data was 14:14 (or 1:1). These results indicate that S. decipiens and S. ranarum are sympatrically distributed in Korea.
-
Key words: Spirometra decipiens, S. ranarum, animals, sympatric distribution, molecular identification, Korea
INTRODUCTION
Species of the genus Spirometra belong to the family Diphyllo-bothriidae and includes intestinal parasites of cats and dogs. These parasites require 2 different intermediate hosts, larval forms of the first intermediate hosts are found in copepods (procercoid) and amphibians and reptiles (plerocercoid) as the second intermediate hosts. Sparganosis or human infection is a zoonotic disease caused by infection with the larval stages of Spirometra species.
The genus
Spirometra has been described with morphological features of spirometrid species under the generic name
Diphyllobothrium as found in China with complex life cycles and include
S. erinaceieuropaei (Rudolphi, 1819),
S. decipiens (Diesing, 1850),
S. ranarum (Gastaldi, 1854),
S. mansoni (Cobbold, 1882)
S. houghtoni (Syn.
S. mansoni, Faust et al., 1929) and
S. okumurai (Faust et al., 1929) by Faust et al. [
1].
Spirometra species in North America have been recognized as
S. mansonoides (Mueller, 1935), which have a characteristic C-shaped outer loop of the uterus [
2]. Five
Spirometra species,
S. decipiens, S. mansoni, S. gracilis (Baer, 1927),
S. longicollis (Parodi and Widakowich, 1917) and
S. mansonoides have been reported from wild fields in South America [
3]. Four
Spirometra species,
S. erinaceieuropaei,
S. pretoriensis (Baer, 1924),
S. theileri (1924) and
S. mansonoides have been acknowledged as valid species by Kamo [
4].
The taxonomy of
Spirometra species has been controversial despite the medical and veterinary importance. Currently, only a few
Spirometra species are considered valid species in the genus
Spirometra. The
Spirometra species currently recognized by many researches worldwide are
S. erinaceieuropaei, S. decipiens, S. mansoni, S. ranarum and
S. mansonoides [
1–
4]. Additionally,
sparganum proliferum is still an unnamed taxon [
5]. A recent report has suggested that there are at least 2
Spirometra species in South America that differ from
S. erinaceieuropaei and
sparganum proliferum [
5]. Unidentified mitochondrial genotypes of
Spirometra species were reported from South Sudan and Ethiopia, in which 37 cases of human sparganosis differed from Asian and South American cases by analysis of mitochondrial DNA sequence data [
6,
7]. The molecular data of
Spirometra species showed that at least 4
Spirometra species such as
S. erinaceieuropaei,
S. decipiens,
S. mansonoides and
sparganum proliferum are distributed in Asian, South American and African countries [
3–
6].
The most recent studies reported identification of
S. ranarum from frogs (
Hoplobatrachus rugulosus; syn:
Rana rugulosa) in Myanmar by morphological and genetic analyses [
8]. Another report demonstrated the distribution of
S. ranarum from lions in Tanzania by analysis of 2 complete mitochondrial genes and morphological observations (to be published).
S. ranarum was first reported by Gastaldi (1854) from
Rana esculenta (syn:
Pelophylax esculentus) in Italy, and Meggitt (1925) described it as
S. ranarum from a dog fed spargana isolated from the same frog host by Gastaldi (1854) in Myanmar [
9,
10]. Following this, Joyeux et al. [
11] and Faust et al. [
1] described
S. ranarum. Wardle and McLeod (1952) recognized
S. ranarum as a valid species [
12]. This
Spirometra species has not been reported since 1929. Currently, mitochondrial DNA sequence evidence combined with examination of morphological features strongly supports the distinctiveness of
Spirometra species, thus the resurrection of
S. ranarum has been proposed in recent reports of
Spirometra species collected from Myanmar and Tanzania (to be published).
The
Spirometra species in the 50 cases of human sparganosis were identified as
S. erinaceieuropaei and
S. decipiens by molecular and morphological features [
3]. Another study identified
S. decipiens plerocercoids (n=904) in terrestrial snakes from Korea and China [
13]. A report concerning the examination of
Spirometra species from a stray cat identified multiple infections of
S. decipiens [
15]. The recent studies suggested that
S. erinaceieuropaei is not the only species inducing human sparganosis but that
S. decipiens is another cause of human sparganosis in Korea [
3,
13,
14].
In the present study, Spirometra species obtained from animals in Korea were identified by molecular analysis of the mitochondrial cytochrome c oxidase I (cox1) gene and phylogenetic analysis of mitochondrial DNA sequence data.
MATERIALS AND METHODS
Specimens
A total of 28
Spirometra species were analyzed in this study (
Table 1). These specimens were collected between 1973 and 2008 in the Republic of Korea. All specimens originated from Korea and obtained from the Department of Parasitology, Gyeongsang National University, Hallym University and Seoul National University. Eight specimens from Gyeongsang National University were collected from a snake (
Rhabdophis tigrinus tigrinus), tadpole and duck were used to infect cats for maintaining the complete life cycle of
Spirometra species in the laboratory. Twelve specimens from Seoul National University were collected from naturally infected cats. Seven specimens from Hallym University were collected from naturally infected dogs. One specimen was collected from leopard cat (
Prionailurus bengalensis), which was donated from the Parasite Resource Bank. Twenty specimens were preserved in 10% neutral buffered formalin, and 8 specimens were kept in 70% ethanol for experimental use.
Total genomic DNA extraction and PCR reactions were employed as previously described by Jeon et al. [
3]. The partial
cox1 gene was amplified and sequenced by PCR and cycle sequencing. The partial sequence of the mitochondrial
cox1 gene was amplified using forward primer p1f, 5′-TGG TTT TTT GGA CAT CCT GAA -3′, and reverse primer p1r, 5′-ATC ACA TAA TGA AAG TGA GCC-3′, which amplified a 440-bp product. A second set of PCR primers was used for cycle sequencing of the internal forward primer p1f1, 5′-GTG TTG ATT TTG CCT GGG TTT-3′, and internal reverse primer p1r1, 5′-TAC AAA CCA AGT ATC ATG TAA-3′, which yielded a 390-bp product. These primers were designed from the complete sequence of
S. erinaceieuropaei (KJ599680) and
S. decipiens (KJ599679) mitochondrial genomes to amplify a partial sequence of the
cox1 gene corresponding to the region between base pair positions 707 and 1,146. The mitochondrial large subunit RNA was amplified using forward primer rRNA F, 5′GAT TTT GTA AAT CAG GGG GTA-3′, and reverse primer rRNA R, 5′-AAT TTA TGC GAT TCA CCT TAA-3′ which amplified a 987 bp product. DNA sequencing was performed using a Big-Dye Terminator kit (version 3.1, Applied Biosystems, Foster City, California, USA) and reaction products were sequenced directly using a DNA sequencer (ABI3730XL, Applied Biosystems).
The DNA sequence of 28 partial
cox1 gene sequences were assembled using the Geneious 9.0 program (Biometer, Auckland, New Zealand) and then aligned using MAFFT methods in the Geneious 9.0 program by comparison with sequences of
S. erinaceieuropaei and
S. decipiens in the GenBank database. Phylogenetic relationships were reconstructed using Bayesian inference (BI) and maximum-likelihood (ML) using partial mitochondrial
cox1 (390 bp) sequences of
S. erinaceieuropaei (KJ599680),
S. decipiens (KJ599679) and
S. ranarum (MH298843). BI analyses were conducted using MrBayes 3.2 and running 4 simultaneous Monte Carlo Markov chains (MCMC) for 10 million generations, sampling every, 1,000 generations and discarding the first 25% generations as burn-in [
15]. BI analysis was evaluated as posterior probability (PP). ML analyses of
cox1 used RAxML v. 7.3.1 [
16] after TRN+G+I substitution model sampling was chosen according to the Modeltest using the program Partition Finder [
17]. Phylogenetic trees were constructed using Bayesian inference (BI) and maximum likelihood (ML) with
Diphyllobothrium nihonkaiense (EF420138) and
D. latum (DQ985706) as outgroups.
RESULTS
Sequence divergences
The mitochondrial
cox1 sequences obtained from Korean isolates of
Spirometra species were compared with the reference
cox1 sequences of
S. erinaceieuropaei,
S. decipiens and
S. ranarum which were deposited in GenBank (accession number KJ599680, KJ5 99679 and MH298843). The mitochondrial
cox1 sequences for a total of 28 specimens were identified as 14
S. decipiens and 14
S. ranarum. The difference in partial
cox1 sequences (316 bp) between
S. erinaceieuropaei (KJ599680) and
S. ranarum (this study) was 9.3%, while that of
S. decipiens (KJ599679) and
S. ranarum (this study) was 2.2%. The sequence identities determined of
Spirometra specimens in this study were 99.8% (
S. ranarum, MH298843), 89.7% (
S. erinaceieuropaei), and 89.7% (
S. decipiens). The similarity to other
Diphyllobothrium species was 84.1% (
D. nihonkaiense) and 83.1% (
D. latum). The similarity of mitochondrial large subunit RNA sequences (987 bp) from Korean isolates to the references sequences was 98.2% (
S. decipiens), 89.4% (
S. erinaceieuropaei), 79.5% (
D. latum) and 80.0% (
D. nihonkaiense) (
Table 2).
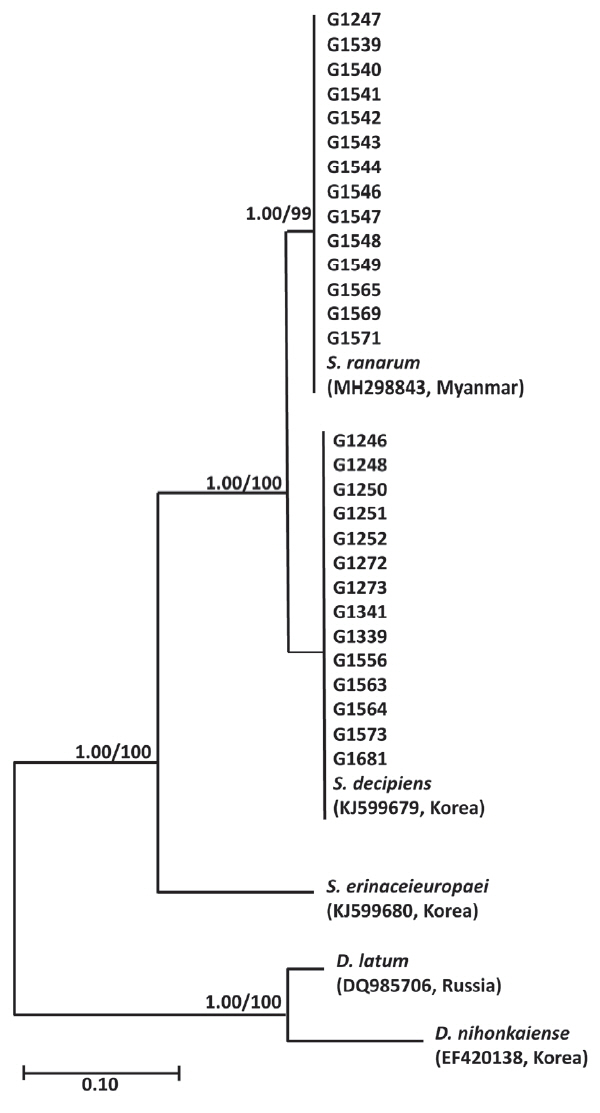
Phylogenetic analyses of
Spirometra species were performed using the Bayesian inference and maximum likelihood methods based on partial mitochondrial
cox1 sequences of
S. erinaceieuropaei,
S. decipiens,
S. ranarum,
D. nihonkaiense and
D. latum. The partial
cox1 sequences (316 bp) revealed 34 polymorphic sites with 34 synonymous and 0 non-synonymous substitutions among
S. erinaceieuropaei,
S. decipiens and
S. ranarum (GenBank no. MH298843). Phylogenetic analysis of the mitochondrial
cox1 sequences for a total of 28 specimens identified
Spirometra species as basal to the
D. nihonkaiense and
D. latum clade. Phylogenetic tree topologies generated using the Bayesian inference and maximum likelihood methods were identical and showed a high level of confidence values for the 3 major branches of the 3
Spirometra species such as
S. erinaceieuropaei,
S. decipiens and
S. ranarum in the
cox1 gene (
Fig. 1).
Genetic analyses identified 2
Spirometra species in wild animals from Korea as
S. decipiens and
S. ranarum.
S. decipiens and
S. ranarum were presented in Gyeongsang, Jeonnam, Gangwon, Chungbuk, and Seoul.
S. decipiens was found in tadpoles, snakes, ducks, cats, leopard cats and dogs while
S. ranarum was found in cats and dogs (
Table 1). The species ratio of
S. decipiens:
S. ranarum calculated from the molecular data was 14:14 (or 1:1) (
Fig. 1).
DISCUSSION
In the present study, we first report
S. ranarum from natural infections of cats and dogs in Korea using mitochondrial
cox1 gene sequence analysis.
S. ranarum (under the name
Ligular ranarum) was first described by Gastaldi (1854) from
Rana esculenta (syn:
Pelophylax esculentus) from Italy. Meggitt (1924) reported the presence of spargana in the stomach wall of frogs (
Rana tigrina) from Yangon, Myanmar. The frogs were found to contain large numbers of a larval tapeworm. These spargana were fed to a young dog and then eight adult tapeworms were recovered 58 days after infection, which the author described as
S. ranarum (under name the
Ligular ranarum) [
9]. Meggitt (1925) described this species and detailed the following features: being up to 1,130 mm in length by 5 mm in breadth, scolex 1.4–1.7 mm in length and 0.37–0.41 mm in breadth, all the segments either broader than long or square, male genital aperture almost at the anterior border of the segment and median, female aperture slightly lateral to it, testes in 2 bands, 100–110 in each band, 3 to 5 uterine coils, uterus extending laterally to the genital apertures, a terminal uterine enlargement, eggs 58–67 by 34–36 μm [
10]. Meggitt et al. [
10] studied the complete life cycle of this species through the intermediate hosts found and showed it to be suitable for final hosts. Faust et al. [
1] studied
S. ranarum (under the name
D. ranarum) from natural infections of cats and dogs in Beijing, Xiamen, Canton and by experimental feeding of spargana obtained from dogs in Fujian.
Spirometra species have been reported sporadically by many authors in the Republic of Korea. Helminth infections such as
Clonorchis sinensis, Paragonimus sp.,
Hydatigera taeniaeformis, Spirometra sp. and
Toxocara cati were examined from 41 cats in Gyeongsangnam-do (Province) [
18]. Seven helminth species,
T. cati, Anisakis simplex larvae,
C. sinensis, Pharyngostomum cordatum, S. erinaceieuropaei and
H. taeniaeformis were reported from 41 cats in Seoul [
19]. Four helminth species including
T. cati, Diphyllobothrium latum, S. erinaceieuropaei and
H. taeniaeformis were detected from 133 cats in Jeollanam-do (Province) [
20]. More than 29 helminth species were reported from feral cats purchased from a market in Busan, and 23 trematodes, 5 cestodes and 4 nematodes species in cats were reported in Korea [
21,
22]. Currently,
S. erinaceieuropaei and
S. decipiens are recognized as being
Spirometra species in Korea [
3]. The first case of human sparganosis in Korea was reported by Uemura [
23]. Snakes and frogs were identified as second intermediate hosts from reports of 63 human sparganosis cases during the years between 1924 and 1974 [
24]. An additional 56 human sparganosis cases were reviewed during the years between 1975 and 1989 [
25].
In this study, we found 2 genotypes in our sequence variation analyses of the cox1 gene from 28 Spirometra specimens obtained from 6 kinds of animals. The sequence difference in the cox1 gene between 14 Spirometra specimens and S. ranarum (GenBank no. MH298843) was 0.1%, while that for the rest of the 14 specimens was 2.2% with S. decipiens and 9.5% with S. erinaceieuropaei. These results indicated that the examined Spirometra specimens in this study were identified as S. decipiens and S. ranarum by mitochondrial DNA sequence divergence. These reports have provoked many questions with respect to the epidemiological discrepancy between humans and animals. In a previous study, human sparganosis cases were identified as S. erinaceieuropaei and S. decipiens, and no cases of S. ranarum were not found in that study. Therefore, although many studies have examined Spirometra species in Korea, those previous studies may need reexamination using molecular techniques to better understand the epidemiological status of Spirometra species in Korea.
The morphological similarity of both adult and larva forms of
Spirometra species have been studied to resolve species identification by use of molecular techniques along with an assessment of morphological variation. Molecular identification has played an important role in improving understanding of phylogenetic relationships, genetic variation and taxonomy. Mitochondrial DNA sequences have been utilized for phylogenetic reconstruction, taxonomic identification, population genetics and epidemiological investigations [
26]. In an effort to delineate the phylogenetic relationships and genetic variation of
Spirometra species, DNA sequence analysis of small (18S) and large (28S) subunit ribosomal RNA, ribosomal internal transcribed spacer 1, ribosomal internal transcribed 2, and mitochondrial genes such as cytochrome c oxidase subunit 1 (
cox1) and 3 (
cox3) and NADH dehydrogenase subunit 1 (
nad1), 3 (
nad3) and 4 (
nad4) have been studied and reported [
27–
32]. Mitochondrial DNA sequence variation of
Spirometra species ranged from 0.0–3.5% in China, Myanmar, Thailand and Lao PDR [
33]. DNA sequence variation of the
Spirometra spp.
cox1 gene ranges from 0.0–2.6% in Japan, India and Indonesia [
34]. The degree of mtDNA sequence divergence of the cytochrome
b (
cob) gene between sister or congeneric species and con-familial genera was greater than 2% in amphibian, reptilian, avian, and mammalian species [
35]. The closely related species of vertebrates showed more than 2% sequence divergence in the
cox1 gene [
36]. Regarding these previous studies, it was assumed that at least 2
Spirometra species were distributed in those endemic areas.
In conclusion, S. decipiens and S. ranarum were identified from natural infections of cats and dogs, with overall results showing 14 S. decipiens and 14 S. ranarum. These results indicate that 2 Spirometra species are sympatrically distributed in Korea.
Notes
-
CONFLICT OF INTEREST
We have no conflict of interest related to this work.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (no. 2017R1D1A3B03035967). Materials used were provided by the Parasite Resource Bank of Korea (PRB000720).
Fig. 1Phylogenetic tree of Spirometra species based on partial cox1 sequences. Numbers above the branches represent bootstrap values for maximum likelihood (ML) and the support values of Bayesian inference (BI) are indicated by the posterior probabilities. S. decipiens and S. ranarum were presented in Gyeongsang, Jeonnam, Gangwon, Chungbuk, and Seoul. S. decipiens was found in tadpoles, snakes, ducks, cats, leopard cats and dogs while S. ranarum was found in cats and dogs. The species ratio of S. decipiens: S. ranarum calculated from the molecular data was 14: 14 (or 1: 1).
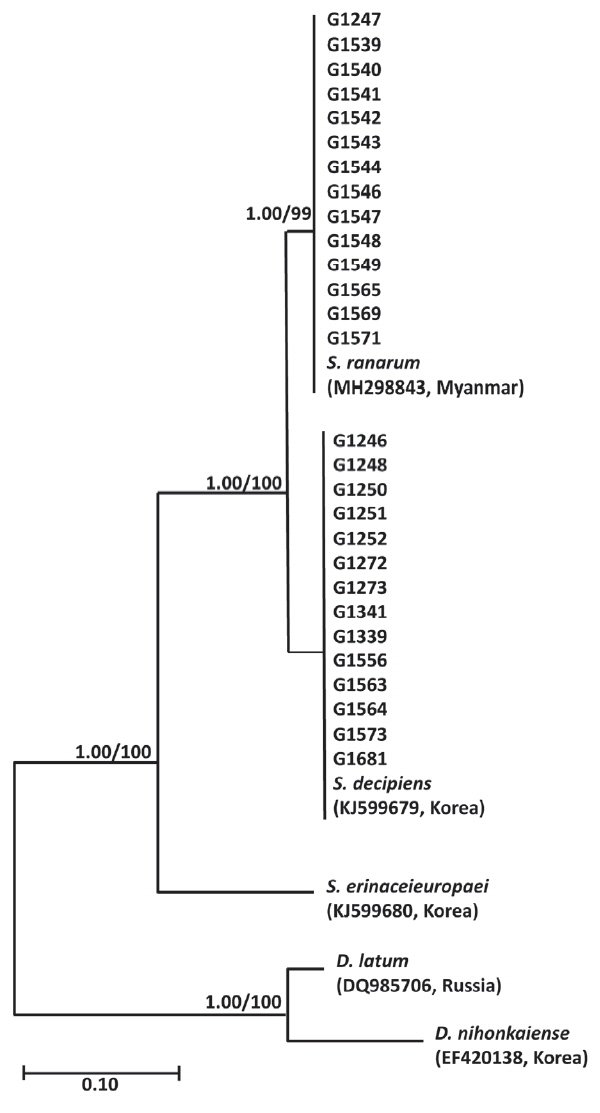
Table 1
Spirometra specimens from animals analyzed in this study (1973–2008)
Table 1
|
Code |
Locality (Korea) |
Host |
Year |
Molecular identification |
|
G1246 |
Jinju |
cat |
2001 |
S.decipiens
|
|
G1247 |
Jinju |
cat |
2001 |
S.ranarum
|
|
G1248 |
Jinju |
cat |
2001 |
S.decipiens
|
|
G1250 |
Jinju |
snake |
2001 |
S.decipiens
|
|
G1251 |
Jinju |
snake |
2001 |
S.decipiens
|
|
G1252 |
Jinju |
tadpole |
2001 |
S.decipiens
|
|
G1272 |
Jinju |
cat |
2001 |
S.decipiens
|
|
G1273 |
Jinju |
duck |
2001 |
S.decipiens
|
|
G1341 |
Seoul |
cat |
1973 |
S.decipiens
|
|
G1339 |
Seoul |
cat |
1987 |
S.decipiens
|
|
G1539 |
Shinan-gun |
cat |
2004 |
S.ranarum
|
|
G1540 |
Shinan-gun |
cat |
2004 |
S.ranarum
|
|
G1541 |
Shinan-gun |
cat |
2004 |
S.ranarum
|
|
G1542 |
Shinan-gun |
cat |
2004 |
S.ranarum
|
|
G1543 |
Shinan-gun |
cat |
2004 |
S.ranarum
|
|
G1544 |
Shinan-gun |
cat |
2004 |
S.ranarum
|
|
G1546 |
Shinan-gun |
cat |
2004 |
S.ranarum
|
|
G1547 |
Shinan-gun |
cat |
2004 |
S.ranarum
|
|
G1548 |
Shinan-gun |
cat |
2004 |
S.ranarum
|
|
G1549 |
Shinan-gun |
cat |
2004 |
S.ranarum
|
|
G1556 |
Chuncheon |
cat |
1988 |
S.decipiens
|
|
G1563 |
Chuncheon |
dog |
2005 |
S.decipiens
|
|
G1564 |
Chuncheon |
dog |
2002 |
S.decipiens
|
|
G1565 |
Chuncheon |
dog |
2002 |
S.ranarum
|
|
G1569 |
Chuncheon |
dog |
1995 |
S.ranarum
|
|
G1571 |
Chuncheon |
dog |
1999 |
S.ranarum
|
|
G1573 |
Chuncheon |
dog |
2000 |
S.decipiens
|
|
G1681 |
Seoul |
leopard cat*
|
2008 |
S.decipiens
|
Table 2Percentage pairwise sequence homologies of the mitochondrial cox1 gene and large subunit ribosomal RNAs between Spirometa sp. of Korea and various Spirometra species, Diphyllobothirum latum and D. nihonkaiense
Table 2
|
Species |
S. ranarum
|
S. decipiens
|
S. erinaceieuropaei
|
D. latum
|
D. nihonkaiense
|
|
GenBank No. |
(MH298843) |
(KJ599679) |
(KJ599680) |
(DQ985706) |
(EF420138) |
|
Genes |
cox1/rRNA |
cox1/rRNA |
cox1/rRNA |
cox1/rRNA |
cox1/rRNA |
|
Spirometra sp. (Korea) |
99.7/100 |
89.7/98.2 |
89.7/89.4 |
83.1/79.5 |
84.1/80.0 |
References
- 1. Faust EC, Campbell HE, Kellogg CR. Morphological and biological studies on the species of Diphyllobothrium in China. Am J Epidemiol 1929;9:560-583.
- 2. Muller JF. New host records for Diphyllobothrium mansonoides Mueller, 1935. J Parasitol 1937;23:313-315.
- 3. Jeon HK, Park H, Lee D, Choe S, Kim KH, Huh S, Sohn WM, Chai JY, Eom KS. Human infections with Spirometra decipiens plerocercoids identified by morphologic and genetic analyses in Korea. Korean J Parasitol 2015;53:299-305.
- 4. Kamo H. Guide to Identification of diphyllobothriid cestodes. Tokyo, Japan. Gendai Kikaku; 1999, pp 1-146 (in Japanese).
- 5. Miyadera H, Kokaze A, Kuramochi T, Kita K, Machinami R, Noya O, Alarcón de Noya B, Okamoto M, Kojima S. Phylogenetic identification of Sparganum proliferum as a pseudophyllidean cestode by the sequence analyses on mitochondrial COI and nuclear sdhB genes. Parasitol Int 2001;50:93-104.
- 6. Almeida GG, Coscarelli D, Melo MN, Melo AL, Pinto HA. Molecular identification of Spirometra spp. (Cestoda: Diphyllobothriidae) in some wild animals from Brazil. Parasitol Int 2016;65:364.
- 7. Eberhard ML, Thiele EA, Yembo GE, Yibi MS, Cama VA, Ruiz-Tiben E. Case report: Thirty-seven human cases of sparganosis from Ethiopia and South Sudan caused by Spirometra spp. Am J Trop Med Hyg 2015;93:350-355.
- 8. Jeon HK, Park H, Lee D, Choe S, Kang Y, Bia MM, Lee SH, Sohn WM, Hong SJ, Chai JY, Eom KS. Genetic and Morphologic Identification of Spirometra ranarum in Myanmar. Korean J Parasitol 2018;56:275-280.
- 9. Meggitt FJ. On the occurrence of Ligular ranarum in a frog. J Nat Hist 1924;9:216-219.
- 10. Meggitt FJ. On the life history of an amphibian tapeworm (Diphyllobothrium ranarum, Gastaldi). J Nat Hist 1925;16:654-655.
- 11. Joyeux C, Baer JG. Sur quelques larvaes de Bothricephales. Bull Soc Path Exot 1927;20:921-936.
- 12. Wardle RA, McLeod JA. The Zoology of Tapeworms. Minneapolis, USA. University of Minnesota Press; 1952, pp 559-615.
- 13. Jeon HK, Park H, Lee D, Choe S, Kim KH, Sohn WM, Eom KS. Genetic identification of Spirometra decipiens plerocercoids in terrestrial snakes from Korea and China. Korean J Parasitol 2016;54:181-185.
- 14. Jeon HK, Park H, Lee D, Choe S, Eom KS.
Spirometra decipiens (Cestoda: Diphyllobothriidae) collected in a heavily infected stray cat from the Republic of Korea. Korean J Parasitol 2018;56:87-91.
- 15. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003;19:1572-1574.
- 16. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006;22:2688-2690.
- 17. Lanfear R, Calcott B, Ho SY, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Bio Evol 2012;29:1695-1701.
- 18. Kang HJ. Studies on the parasitic helminths of the cats in western province of Kyung Sang Nam-do. Res Bull Chinju Agric Coll 1967;6:91-96. (in Korean).
- 19. Huh S, Sohn WM, Chai JY. Intestinal parasites of cats purchased in Seoul. Korean J Parasitol 1993;31:371-373.
- 20. Yang HJ, Park TW, Cheon SJ, Yoon YB, Kim NJ, Park BK, Kim CS. Internal parasites of cats in Iri and its vicinity. Korean J Vet Serv 1995;18:33-40. (in Korean).
- 21. Sohn WM, Chai JY. Infection status with helminthes in feral cats purchased from a market in Busan, Republic of Korea. Korean J Parasitol 2005;43:93-100.
- 22. Chai JY, Bahk YY, Sohn WM. Trematodes recovered in the small intestine of stray cats in the Republic of Korea. Korean J Parasitol 2013;51:99-106.
- 23. Uemura S. On the Ligula mansoni from human. J Chosen Med Ass 1917;20:114. (in Japanese).
- 24. Cho SY, Bae JH, Seo BS. Some aspects of human sparganosis in Korea. Korean J Parasitol 1975;13:60-77.
- 25. Min DY. Cestode infections in Korea. Korean J Parasitol 1990;28(suppl):123-144.
- 26. Le TH, Blair D, McManus DP. Mitochondrial genomes of parasitic flatworms. Trends Parasitol 2002;18:206-213.
- 27. Dai RS, Liu GH, Song HQ, Lin RQ, Yuan ZG, Li MW, Huang SY, Liu W, Zhu XQ. Sequence variability in two mitochondrial DNA regions and internal transcribed spacer among three cestodes infecting animals and humans from China. J Helminthol 86:245-251.
- 28. Liu W, Zhao GH, Tan MY, Zeng DL, Wang KZ, Yuan ZG, Lin RQ, Zhu XQ, Liu Y. Survey of Spirometra erinaceieuropaei spargana infection in the frog Rana nigromaculata of the Hunan Province of China. Vet Parasitol 2010;173:152-156.
- 29. Zhang X, Cui J, Wei T, Li LY, Jiang J, Lu JC, Jiang P, Liu LN, Wang ZQ. Survey and genetic variation of Spirometra erinaceieuropaei sparganum in frogs and snakes from Guangxi of southern China. Trop Biomed 2014;31:862-870.
- 30. Zhang X, Wang H, Cui J, Jiang P, Lin ML, Zhang YL, Liu RD, Wang ZQ. The phylogenetic diversity of Spirometra erinaceieuropaei isolates from southwest China revealed by multi genes. Acta Trop 2016;156:108-114.
- 31. Zhang X, Duan JY, Wang ZQ, Jiang P, Liu RD, Cui J. Using the small subunit of nuclear ribosomal DNA to reveal the phylogenetic position of the plerocercoid larvae of Spirometra tapeworms. Exp Parasitol 2017;175:1-7.
- 32. Zhang X, Duan JY, Shi YL, Jiang P, Zeng DJ, Wang ZQ, Cui J. Comparative mitochondrial genomics among Spirometra (Cestoda: Diphyllobothriidae) and the molecular phylogeny of related tapeworms. Mol Phylogenet Evol 2017;117:75-82.
- 33. Jongthawin J, Intapan PM, Sanpool O, Sadaow L, Laymanivong S, Thanchomnang T, Maleewong W. Molecular evidence of Spirometra erinaceieuropaei infection in snakes Ptyas korros from Lao PDR and Thailand and frogs Hoplobatrachus rugulosus from Myanmar. Southeast Asian J Trop Med Public Health 2014;45:1271-1278.
- 34. Okamoto M, Iseto C, Shibahara T, Sato MO, Wandra T, Craig PS, Ito A. Intraspecific variation of Spirometra erinaceieuropaei and phylogenetic relationship between Spirometra and Diphyllobothrium inferred from mitochondrial CO1 gene sequence. Parasitol Int 2007;56:235-238.
- 35. Johns GC, Avise JC. A comparative summary of genetic distances in the vertebrates from the mitochondrial cytochrome b gene. Mol Bio Evol 1998;15:1481-1490.
- 36. Hebert PD, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci 2003;(suppl):96-99.