Abstract
Recent in vitro studies have revealed that a certain Mycobacterium can survive and multiply within free-living amoebae. It is believed that protozoans function as host cells for the intracellular replication and evasion of Mycobacterium spp. under harmful conditions. In this study, we describe the isolation and characterization of a bacterium naturally observed within an amoeba isolate acquired from a contact lens storage case. The bacterium multiplied within Acanthamoeba, but exerted no cytopathic effects on the amoeba during a 6-year amoebic culture. Trasnmission electron microscopy showed that the bacteria were randomly distributed within the cytoplasm of trophozoites and cysts of Acanthamoeba. On the basis of the results of 18S rRNA gene analysis, the amoeba was identified as A. lugdunensis. A 16S rRNA gene analysis placed this bacterium within the genus Mycobacterium. The bacterium evidenced positive reactivity for acid-fast and fluorescent acid-fast stains. The bacterium was capable of growth on the Middlebrook 7H11-Mycobacterium-specific agar. The identification and characterization of bacterial endosymbionts of free-living protozoa bears significant implications for our understanding of the ecology and the identification of other atypical mycobacterial pathogens.
-
Key words: Acanthamoeba, Mycobacterium, 18S rRNA, endosymbiont
INTRODUCTION
The genus
Acanthamoeba is an amphizoic amoeba, which is a causative agent of vision-threatening keratitis and life-threatening granulomatous encephalitis (
Khan, 2006). One more medically important feature of the genus is that it is well-known to function as a carrier or vehicle for some pathogenic microbes, including
Legionella pneumophila,
Vibrio cholerae, and
Listeria monocytogenes (
Holden et al., 1984;
Ly and Muller, 1990;
Thom et al., 1992). The amoeba host would provide shelter to bacterial species against harmful environments. These interactions enable the bacteria and amoebae to establish an endosymbiotic relationship, which closely resembles the pathogenesis of chronic bacterial infections within mammalian cells (
Ahn et al., 1990;
Park, 1990).
Acanthamoeba has been suspected as an environmental host of
Mycobacterium sp. since Jadin's (
1973) report regarding the existence of acid-fast stained bacteria inside of the amoeba. Akin to the intracellular parasitism of
Legionella (
Kilvington and Price, 1990;
Fields, 1996),
Mycobacterium would multiply within
Acanthamoeba trophozoites and cysts. There have been a host of investigations conducted in an attempt to elucidate the endosymbiotic association between
Acanthamoeba and
Mycobacterium via artificial infection laboratory trials (
Cirillo et al., 1997;
Steinert et al., 1998;
Miltner and Bermudez, 2000;
Taylor et al., 2003). Investigators reported that the coculturing of
L. pneumophila and
M. avium with
Acanthamoeba caused increased virulence in the bacterial and amoebic hosts (
Cirillo et al., 1994,
1997). However, the artificially-induced transient association between the bacterial and amoebic hosts has made any detailed research into the mechanisms underlying the interaction between the endosymbiotic bacteria and
Acanthamoeba difficult.
In the present study, we found an endosymbiotic bacterium of Acanthamoeba during the characterization of environmental Acanthamoeba isolates from various origins via mitochondrial (Mt) DNA RFLP. The Mt DNA RFLP pattern of Acanthamoeba KA/LC6 isolated from contact lens storage case evidenced several extra bands, considering the general size of the Mt DNA of Acanthamoeba. We verified the existence of an endosymbiotic bacterium via transmission electron microscopy and identified this endosymbiotic bacterium as Mycobacterium sp., on the basis of its molecular and biochemical characteristics.
MATERIALS AND METHODS
Acanthamoeba
Acanthamoeba KA/LC6 was isolated from a contact lens storage case in Korea. Two strains, A. castellanii Castellani and A. lugdunensis L3a, were employed as reference strains. The amoeba was cultured in PYG media at 25℃.
Mitochondrial (Mt) DNA RFLP analysis of Acanthamoeba
The MtDNA of
Acanthamoeba KA/LC6 and the reference strains were extracted via the technique developed by Yagita and Endo (
1990). In brief, the MtDNA from amoeba trophozoites was extracted with phenol/ chloroform and precipitated with absolute ethanol and sodium acetate solution.
EcoR I restriction endonuclease (Promega, Madison, Wisconsin, USA) was utilized for MtDNA RFLP analysis. The digested DNA was electrophoresed and stained with ethidium bromide. The MtDNA RFLP patterns of the three strains were observed and photographed under a UV transilluminator.
The trophozoite and cyst suspension was centrifuged for 10 min at 2,000 rpm and the sediment was washed 3 times in cold phosphate-buffered saline (PBS). The sediment was prefixed for 2 hr with 4% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2-7.4. After rinsing with 0.1 M cacodylate buffer, the sediment was post-fixed for 3 hr with 1% osmium tetraoxide, rinsed twice with 0.1 M maleate buffer, pH 5.2, dehydrated with ethyl alcohol, and treated for 30 min with propylene oxide. The pellet was then immersed in propylene oxide-resin (1:1) overnight with continuous shaking. The pellet was embedded in resin and incubated overnight at 60℃. Ultra-thin sections cut with a Reichert-Jung ultramicrotome were then stained with uranyl acetate and lead citrate. The sections were observed under a Hitachi-7000 electron microscope (Hitachi-7000, Tokyo, Japan).
AFB (acid fast bacilli) and fluorescent AFB staining
Diluted Acanthamoeba KA/LC6 or A. lugdunensis L3a in PBS were spotted onto a slide glass and dried for 5 min at 70℃. For the AFB staining, the slides were incubated on Siehl-Neelsen carbofuchsin stain solution for 5 min with heat. Slides washed with distilled water were incubated in acid alcohol for 15-20 sec. The slides were again washed with distilled water and incubated in Loeffler methylene blue stain solution for 30 sec. Air-dried slides were examined under lightmicroscopy. For FAFB, the slides were flooded with an auramine O-rhodamine B-phenol stain solution (1.5 g of auramine O (Sigma-Aldrich Co., St. Louis, USA), 0.75 g of rhodamine B (Matheson Coleman and Bell, Norwood, USA), 75 ml of glycerol, 10 ml of phenol, and 50 ml of distilled water) and allowed to stain for 15 minutes. The slides were rinsed in chlorine-free water and flooded with 0.5% acid alcohol (0.5 ml of concentrated HCl in 100 ml of 70% ethanol) in order to decolorize for 2 min. The slides were then rinsed with chlorine-free water and incubated for 3 min in potassium permanganate solution. The slides were read on a fluorescent microscope.
PCR-based amplification of 16S rDNA of endosymbiont and 18S rDNA of Acanthamoeba
The PCR-amplification of the 16S rDNA of the bacterial endosymbiont was conducted with the Mt DNA of
Acanthamoeba KA/LC6 and two reference strains. The sequence of eubacterium-specific 16s rDNA primers was as follows: 5'-CCG AAT TCG TCG ACA ACA GAG TTT GAT CCT GGC TCA G-3' and 5'-CCC GGG ATC CAA GCT TAA GGA GGT GAT CCA GCC-3'. For the PCR amplification of
Acanthamoeba 18S rDNA, we employed the genomic DNA of
Acanthamoeba, which was obtained via the method described by Kong and Chung (
1996), as templates, and the following primers: 5'-CCG AAT TCG TCG ACA ACC TGG TTG ATC CTG CCA GT-3' and 5'-GGA TCC AAG CTT GAT CCT TCT GCA GGT TCA CCT AC-3', which were designed by Chung et al. (
1998). Each reaction mixture contained 0.1-1.0 ng DNA template, 1.25 U
Taq polymerase (SUPER BIO, Suwon, Korea), 200 M dNTPs, 1.5 mM MgCl
2 and 0.5 M of each primer. The reaction mixtures were then brought to a volume of 50 µl with sterile distilled water. The cycling of the program progressed as follows: an initial 5 min at 94℃ followed by 30 cycles of 94℃ 20 sec, 60℃ for 30 sec, 72℃ for 1.5 min, and a final 10-min elongation step at 72℃. In the case of the
Acanthamoeba 18S rDNA, we conducted a 2.5-min DNA elongation step.
The amplified DNA of the 16S rDNA of the endosymbiont and the 18S
Acanthamoeba rDNA were purified via electrophoresis in low-melting-point agarose and ligated into the cloning vector pGEM
®-T easy Vector System I (Promega). Each vector was subsequently transformed into into
E. coli. The vector DNA prepared via plasmid DNA extraction was sent to Macrogen (Macrogen, Seoul, Korea) for sequence analysis. A BLAST search of the 16S rDNA sequences of the endosymbionts of
Acanthamoeba KA/LC6 through a public database yielded matching 16S rDNA sequences in 19
Mycobacterium spp. We aligned the sequences of the endosymbiont and closely related mycobacterial species using the CLUSTALX program. The phylogenetic tree was constructed via the unweighted pair group method with arithmetic average (UPGMA), using the Phylip program, version 3.5 (
Felsenstein, 1993).
In order to determine the growth of endosymbiotic bacteria on a cell-free medium, bacteria freed from the amoeba host were incubated on Mycobacterium specific media, Middlebrook 7H11 agar. The trophozoites of Acanthamoeba KA/LC6 (approximately 1 × 105) were washed 3 times with PBS and sonicated for 0, 1, 5, 10, 30, 60, or 90 sec. The sonicated cell suspensions were inoculated on Middlebrook 7H11 agar and cultured in an incubator at 37℃. The culture tubes were monitored for bacterial growth every week for 1 mo.
RESULTS
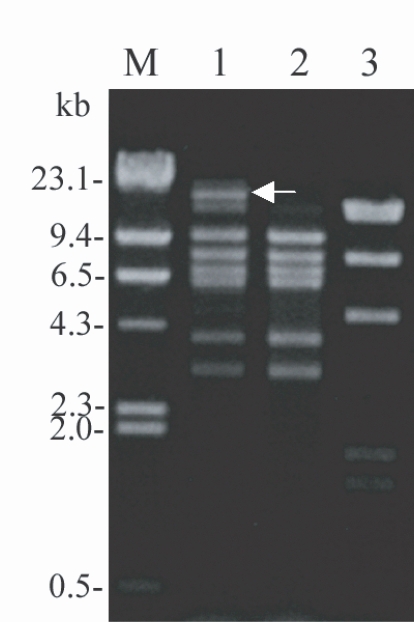
Extra-fragments in MtDNA RFLP of Acanthamoeba KA/LC6
The MtDNA RFLP pattern of the
Acanthamoeba KA/LC6 isolated from a contact lens storage case was almost identical to that of
A. lugdunensis L3a, with the exception of several extra fragments (
Fig. 1). This was very different from the pattern of
A. castellanii Castellani. The rough sum of all DNA fragment sizes of KA/LC6 was significantly more than 45 kb, the average size of
Acanthamoeba mitochondrial DNA (
Burger et al., 1995). This is suggestive of the presence of extra chromosomal DNA, such as plasmid DNA, from the bacterial endosymbiont.
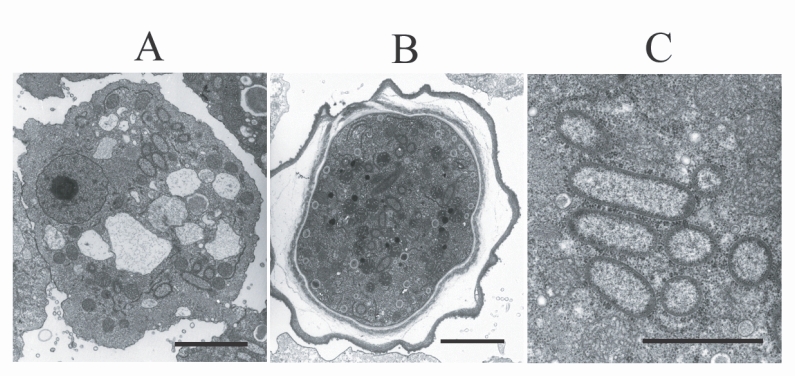
TEM examination revealed the presence of endosymbiotic bacteria within the amoeba (
Fig. 2). The bacteria were observed in both the trophozoites and cysts, and were found randomly distributed throughout the amoebic cytoplasm. The endosymbiont, which was sized 1.40 × 0.49 µm, was a double-membraned rod-shaped bacterium, studded on its surface with host ribosomes. No lacunae or vesicle-like structures were detected around the bacterium.
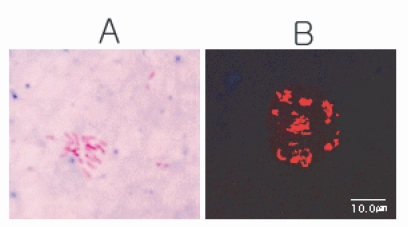
The acid-fast stained
Acanthamoeba harbored rod-shaped bacteria within the trophozoite and the cyst (
Fig. 3A). The bacteria were approximately 1.5 µm in length. A fluorescent AFB stain specific for
Mycobacterium verified the presence of
Mycobacterium within the amoebic trophozoite. A number of red-colored, rod-shaped bacteria were scattered in groups within the amoebic cytoplasm (
Fig. 3B).
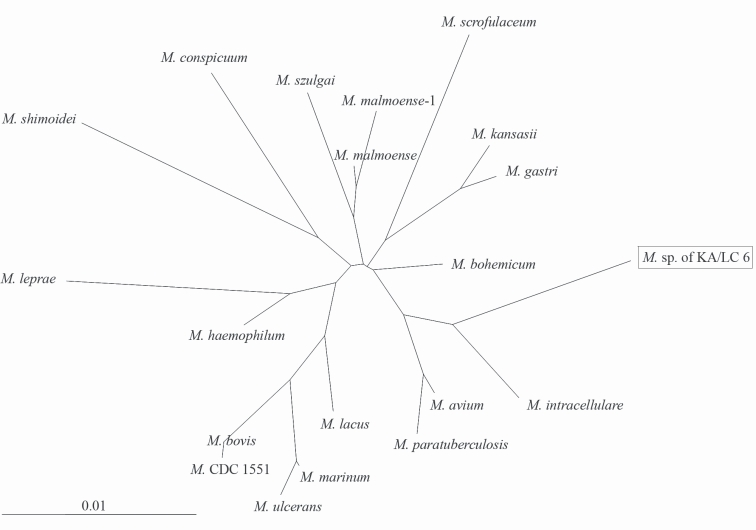
Via a homology search using a public database, the 16S rDNA sequence of the endosymbiont was determined to have a profound DNA similarity with those of
Mycobacterium species. In the phylogenetic tree (
Fig. 4) constructed on the basis of results of the 16S rDNA sequence analyses of 19
Mycobacterium spp. including the present isolate, the endosymbiont of
Acanthamoeba KA/LC6 was determined to be related most closely to
M. avium and
M. intracellulare. The host
Acanthamoeba was identified as
A. lugdunensis via 18s rDNA sequence analysis and its MtDNA RFLP pattern.
Yellow colonies were observed after one month in all tubes under a variety of sonication conditions. Interestingly, the largest numbers of colonies were observed in the culture tube with the unsonicated amoeba, thereby indicating either that Mycobacterium could be freed from Acanthamoeba spontaneously, or that the sonication had harmed the Mycobacterium samples.
DISCUSSION
In this study, we have identified and characterized an endosymbiotic bacterium of
Acanthamoeba, which was isolated from a contact lens storage case. The endosymbiotic bacterium of
Acanthamoeba KA/LC6 evidenced 98% 16s rRNA gene sequence similarity with that of
Mycobacterium sp. This is the first genetically verified natural occurrence of
Mycobacterium in
Acanthamoeba. The endosymbiotic
Mycobacterium sp. was found to be closely genetically related to the
M.avium complex, which is generally considered to be a very important causative agent of tuberculosis in immunocompromised hosts, most notably AIDS patients (
Reed et al., 2006).
It has been suspected that
Mycobacterium might be transferred by free-living amoeba, including
Acanthamoeba (
Winiecka-Krusnell and Linder, 2001). Many papers have focused on the relationship between
Mycobacterium and
Acanthamoeba in an induced transient endosymbiosis system (
Cirillo et al., 1997;
Steinert et al., 1998;
Miltner and Bermudez, 2000;
Taylor et al., 2003). The location of endocytosed mycobacterium in vacuolar structures, as is observed in cases of efficient escape from the cellular phagocytic/ endocytic degradation system, constitutes a crucial strategy for the survival of intracellular bacteria (
Amano et al., 2006). Although the exact mechanism underlying this phenomenon remains to be fully understood,
Mycobacterium is known to inhibit phagosome-lysosome fusion, allowing it to survive within human macrophages (
Smith, 2003). A similar phenomenon was observed in the artificially-induced symbiosis of
Acanthamoeba and
Mycobacterium. In an in vitro study,
M. avium was found to inhibit lysosomal fusion and to replicate in vacuoles that are in a tight juxtaposition with the bacterial surface within amoebae (
Cirrillo et al., 1997). The morphological characteristics of the endosymbiotic
Mycobacterium presented in this paper, however, are quite different from those of the
Mycobacterium in
Acanthamoeba hosts induced by in vitro coincubation. The
Mycobacterium in
Acanthamoeba KA/LC6 was not closed by any membraneous structure, and somehow escaped the vesicles and became dispersed within the cytoplasm of the amoebic host. Some intracellular bacteria escape from the intracellular endocytic compartment to the cytoplasm in order to circumvent or elude the host degradation system.
Listeria monocytogenes normally enters host cells via endocytosis and escapes from the intracellular endosomes (
Amer and Swanson, 2002;
Rosenberger and Finlay, 2003). An investigation of the mechanism underlying the escape from the endocytic compartment to the cytoplasm would be a good direction for future research into the relationship between
Mycobacterium sp. and
Acanthamoeba KA/LC6.
The host,
Acanthamoeba KA/LC6, was identified as
A. lugdunensis via 18S rDNA sequence analysis. The MtDNA RFLP patterns were identical to those of the L3a strain, the type strain of
A. lugdunensis. This L3a genotype was most frequently isolated from the corneas of keratitis patients, and from environmental samples, including contact lens storage cases and hospital water supplies in Korea (
Shin et al., 1999;
Yu et al., 2004). Two
Acanthamoeba isolates, one obtained from a contact lens storage case and the other acquired from a water cooling tower in Korea, were reported to contain endosymbiotic bacteria (
Kong and Chung, 1996;
Chung et al., 1997). The two isolates also evidenced the L3a genotype. There is as yet no direct evidence that the L3a genotype has better biological conditions for the establishment of bacterial symbiosis. Genomic and proteomic approaches to determine the differences between L3a and the other genotypes might provide the necessary clues to understand the mechanisms relevant to symbiosis.
The transient association of
Mycobacterium and
Acanthamoeba by coculture has limitations with regard to the investigation of long-term genetic influences on the gene expression and protein profiles of both the bacteria and the
Acanthamoeba host. Symbionts may alter the host's gene expression of an essential protein. In the case of X-bacteria and
Amoeba proteus, long-term symbiosis resulted in an alteration of the protein profile of the amoebic host (
Choi et al., 1997). X-bacteria induced irreversible nucleolar abnormalities to the amoeba host in order to stop the production of S-adenosylmethionine synthetase permanently. However, little remains known regarding the molecular and biochemical alterations inherent to the association of endosymbionts and the
Acanthamoeba host. The naturally-occurring endosymbiosis and the isogenetic partner of
Acanthamoeba-KA/L6 and L3a- would constitute a good model system for studies of long-term genetic influences on the gene expression and protein profiles of both the bacteria and the amoebic host.
Notes
-
This research was supported by Kyungpook National University Research Team Fund, 2002.
References
Fig. 1Agarose gel electrophoretic restriction fragment patterns for the mitochondrial DNA of Acanthamoeba KA/LC6 and the reference strains. Lane 1: Acanthamoeba KA/LC6, 2: A. lugdunensis L3a, 3: A. castellanii Castellani. M: Hind III digested λ phage DNA used as a DNA size standard. Arrow indicates extra fragments in comparison with the reference strains.

Fig. 2Electron micrographs of
Acanthamoeba KA/LC6 with bacterial endosymbionts.
A. Rod-shaped bacterial endosymbionts are randomly distributed throughout the trophozoitic cytoplasm.
B. Cyst with endosymbionts.
C. Magnification of
Fig. 2A. Ribosomes of the amoebic host are studded on the surfaces of the endosymbionts. Bar indicates 2 µm.
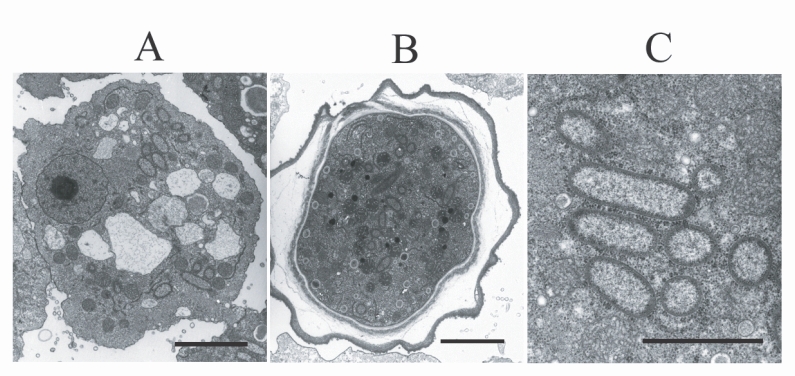
Fig. 3Stained endosymbionts in Acanthamoeba KA/LC6. A. An Acanthamoeba KA/LC6 harbors numerous acid-fast, red-staining, rod-shaped mycobacterium stained with Ziehl-Neelsen stain. B. Fluorescent AFB stain specific for mycobacterium revealed many rod-shaped bacteria in Acanthamoeba KA/LC6.

Fig. 4Neighbor-joining dendrogram based on the results of comparative 16S rRNA analysis, showing the relationships of the endosymbiont of Acanthamoeba KA/LC6 and M. bohemicum to representative members of the genus Mycobacterium (bar represents 1% estimated evolutionary distance).