Abstract
In the present study, genomic DNAs were purified from Korean isolates (KT8, KT6, KT-Kim and KT-Lee) and foreign strains (CDC85, IR78 and NYH 286) of Trichomonas vaginalis, and hybridized with a probe based on the repetitive sequence cloned from T. vaginalis to observe the genetic differences. By Southern hybridization, all isolates of T. vaginalis except the NYH286 strain had 11 bands. Therefore all isolates examined were distinguishable into 3 groups according to their banding patterns; i) KT8, KT6 and KT-Kim isolates had 11 identical bands such as 1 kb, 1.2 kb, 1.6 kb, 1.9 kb, 2.3 kb, 2.7 kb, 3.2 kb, 3.4 kb, 3.8 kb, 4.9 kb and 6.0 kb. ii) The metronidazole-resistant IR78 strain had the same bands as KT-Lee isolate at bands of 1 kb, 1.2 kb, 1.6 kb, 1.8 kb, 2.1 kb, 2.5 kb, 2.7 kb, 2.9 kb, 3.4 kb, 5.0 kb and 6.0 kb. Bands of CDC85, metronidazole-resistant strain, were similar to those of IR78 and KT-Lee, except that 3.2 kb replaced 2.9 kb. iii) NYH286 particularly had 12 bands and band patterns were similar to IR78 with a few exceptions as follows: i) 6.2 kb in place of 6.0 kb, ii) 2.0 kb and 2.2 kb instead of 2.1 kb. Through the results obtained, genetic variance of T. vaginalis isolates was demonstrated by Southern hybridization.
-
Key words: Trichomonas vaginalis, Southern hybridization, genetic diversity
Trichomonas vaginalis is one of the most widespread sexually transmitted diseases in the world with an incidence of 10 to 50% in patients of sexually transmitted disease clinics (
Spence, 1986).
The Southern blot analysis has been introduced for distinguishing species of
Giardia isolate, differentiating the human infective and non-human infective isolates of
Trypanosoma brucei and identification of
Leishmania strains and
T. vaginalis (
Nash et al., 1985;
Rubino et al., 1991;
Guizani et al., 1994;
Hide et al., 1994), In this study, the genetic variance of
T. vaginalis were examined by Southern hybridization.
Seven isolates of
T. vaginalis and one isolate of
Tritrichomonas foetus were used in the study.
Trichomonas vaginalis KT8, KT6, KT-Kim and KT-Lee isolates were obtained from Korean women with acute vaginitis (
Ryu et al, 1995). Metronidazole resistant
T. vaginalis CDC85, IR78 and
T. foetus KV strain were kindly provided by professor David Lloyd from the School of Pure and Applied Biology, Wales University, Cardiff. The NYH286 strain was purchased from ATCC (USA). Trophozoites were axenically cultured in TYM medium (
Diamond, 1957).
For genetic analysis, it is necessary to get homogenous clone. The cloning of
T. vaginalis was done according to the method applied to
Entamoeba histolytica with a modification as 10% CO
2 incubator instead of anaerobic jar (
Mueller and Petri, 1995). The agar (5%) and trophozoites (1 × 10
3) were mixed by inverting the tube 10 times and contents were then Immediately poured into a Petri dish (100 by 15 mm). The colonies were visible to the naked eye after 4-5 days. Among seven isolates, KT8, KT-Lee and IR78 isolates had 2 colonies and CDC85 showed one colony (
Fig. 1). We named the colony of each isolates as KT8C1 and KT8C2, KT-LeeCl and KT-LeeC2, IR78C1 and IR78C2 and CDC85C, and cultivated each colony separately.
Trichomonad genomic DNA was isolated by a modified method of Rubino et al. (
1991) and Ho et al (
1994). Briefly, cells were lysed with a NET buffer (containing 0.2% SDS) and DNA was extracted with phenohchloroform, precipitated with sodium acetate, rinsed with ethanol, and dissolved in a TE buffer. RNase and proteinase K were treated subsequently, and then DNA was extracted with phenol: chloroform. The following procedures were the same as described above.
Field inversion gel electrophoresis (FIGE) was done using FIGE mapper (Bio-Rad). Five µg of
EcoRl digested genomic DNA was loaded to each lane. Running condition was set at program 1 for 8 hr, and program 2 for 8 hr, and forward and reverse voltage was 180 V and 120 V, respectively. Restriction fragments separated on 0.8% agarose gels were denatured and transferred onto nylon membrane (Hybond-N+, Amersham) by Southern blotting (
Southern, 1975). For the probe preparation, PCR was done with
T. vaginalis specific repetitive primers designed from TV-E650-1, cloned by Pace et al (
1992). The forward and reverse primers were 5' GAGTTAGGGTCTAATGTTTGATGTG 3' and 5' AGAATGTGATAGCGAAATGGG 3' respectively. The amplification cycle comprised an initial 5 min jdenaturation at 95℃, then 1 min at 94℃, 1 min at 50℃, 1 min at 72℃ for 35 cycle. About 337 bp sized product was identified and purified from agarose gels by using a Gene-clean kit (Qiagen, Germany). The purified 337 bp fragment was labeled with [α-
32P]dCTP by PCR as described previously (
Konat et al, 1994). Hybridization was carried out overnight in a 5 × Denhardt's solution containing 6 × SSPE, 50% formamide. salmon sperm DNA (100 µg/ml), and 0.5% SDS at 42℃. Membranes were washed twice with 2 × SSPE and 0.5 × SSPE. The autoradiographs were exposed to X-Omat film (Kodak, NY) with an intensifying screen for 3 weeks at -70℃.
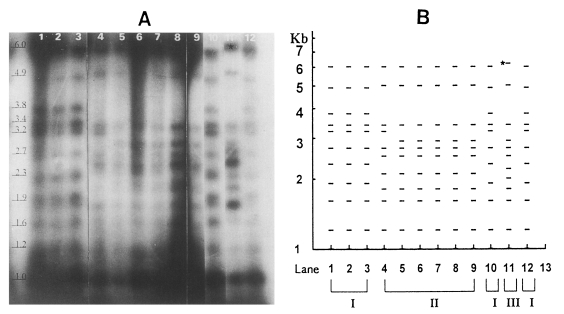
By Southern hybridization, all six isolates of
T. vaginalis except NYH286 strain had 11 bands in contrast to
T. foetus showing no hybridization band (
Fig. 2A). Bands shown by trophozoites of
T. vaginalis before cloning were the same as those of colony-forming trophozoites after cloning. Namely, bands of the KT8 isolate were identical to KT8C1 and KT8C2; the same as in IR78 and KT-Lee (
Fig. 2A).
Three groups of isolates were distinguished after hybridization (
Fig. 2B). The KT8, KT6 and KT-Kim isolates of the first group had 11 identical bands: 1 kb, 1.2 kb, 1.6 kb, 1.9 kb, 2.3 kb, 2.7 kb, 3.2 kb, 3.4 kb, 3.8 kb, 4.9 kb and 6.0 kb. The second group comprised IR78. KT-Lee and CDC85. The metronidazole-resistant IR78 strain had the same bands as the KT-Lee isolate: 1 kb, 1.2 kb, 1.6 kb, 1.8 kb, 2.1 kb, 2.5 kb, 2.7 kb, 2.9 kb, 3.4 kb, 5.0 kb and 6.0 kb. CDC85 is known as metronidazole-resistant strain, IR78. The bands of CDC85 were similar to those of IR78 and KT-Lee. except that 3.2 kb replaced 2.9 kb. The NYH286 strain, the third one, had 12 bands, and its hybridization pattern had shared common bands with IR78 with a few exceptions as follows: i) 6.2 kb in place of 6.0 kb. ii) 2.0 kb and 2.2 kb instead of 2.1 kb (
Fig. 2A). Both strains of IR78 and CDC85 were metronidazole-resistant although metronida-zole-sensitive KT-Lee had bands identical to IR78. Thus, the present hybridization pattern showed no relation with metronidazole resistance.
In the present study we have used EcoRI as a restriction enzyme for digestion of genomic DNA. Another restriction enzyme treatment probably will show band patterns different from bands showed in this experiments. As the seven isolates of T. vaginalis were not enough to observe the genetic variance, we are planning a further research with more isolates of T. vaginalis and restriction enzymes for examining genetic differences of T. vaginalis.
Notes
-
This study was supported by the Research Grant of the Faculty Research Project, Hanyang University, 1996.
ACKNOWLEDGEMENTS
The authors would like to thank Mr. Han-Gyoo CHOI and Mrs. So-Young PARK, Department of Parasitology. College of Medicine, Hanyang University and Mr. Bong-Yoon KIM, Department of Medical Genetics, College of Medicine, Hanyang University for their technical help during the study.
References
- 1. Alderete JF, Kasmala L, Metcalfe E, Garza GE. Phenotypic variation and diversity among Trichomonas vaginalis isolates and correlation of phenotype with trychomonal virulence determinants. Infect Immun 1986;53:285-293.
- 2. Diamond L. The establishment of various trichomonads of animal and man in axenic cultures. J Parasitol 1957;43:488-490.
- 3. Guizani I, Van Eys GJJM, Ben Ismail R, Dellagi K. Use of recombinant DNA probes for species identification of old world Leishmania isolates. Am J Trop Med Hyg 1994;50(5):632-640.
- 4. Hide G, Welburn SC, Tait A, Maudlin I. Epidemiological relationships of Trypanosoma brucei stocks from South East Uganda: evidence for different population structures in human infective and non-human infective isolates. Parasitology 1994;109:95-111.
- 5. Ho MSY, Conrad PA, Conrad PJ, LeFebvre RB, Perez E, BonDurant RH. Detection of bovine trichomoniasis with a specific DNA probe and PCR amplification system. J Clin Microbiol 1994;32:98-104.
- 6. Konat GW, Laszkiewicz I, Grubinska B, Wiggins RC. In Griffin HG, Griffin AM eds, Generation of labeled DNA probes by PCR. PCR technology. Current innovations. 1994, Florida, USA. CRC Press Inc; pp 37-42.
- 7. Mason PR, Forman L. Polymorphonuclear cell chemotaxis to secretions of pathogenic and nonpathogenic Trichomonas vaginalis. J Parasitol 1982;68:457-462.
- 8. Min DY, Ryu JS, Park SY, Shin MH, Cho WY. Degradation of human immunoglobulins and cytotoxicity on HeLa cells by live Trichomonas vaginalis. Korean J Parasitol 1997;35(1):39-46.
- 9. Mueller DE, Petri WA. Clonal growth In petri dishes of Entamoeba histolytica. Trans Royal Soc Trop Med Hyg 1995;89:123.
- 10. Nash TE, McCutchan T, Keister D, Dame JB, Conrad JD, Gillin FD. Restriction-endonuclease analysis of DNA from 15 Giardia isolates obtained from humans and animals. J Infect Dis 1985;152(1):64-73.
- 11. Pace J, Urbankova V, Urbanek P. Cloning and characterization of a repetitive DNA sequence specific for Trichomonas vaginalis. Mol Biochem Parasitol 1992;54:247-256.
- 12. Rubino S, Muresu R, Rappelli P, et al. Molecular probe for identification of Trichomonas vaginalis DNA. J Clin Microbiol 1991;29(4):702-706.
- 13. Ryu JS, Park JW, Min DY. Effect of sodium nitrite on Trichomonas vaginalis. Korean J Parasitol 1995;33(4):349-356.
- 14. Soliman MA, Ackers JP, Catterall . Isoenzyme characterization of Trichomonas vaginalis. Br J Vener Dis 1982;58:250-256.
- 15. Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 1975;98:503-517.
- 16. Spence MR. In Felman TM ed, Gardnerella and Trichomonas vaginitis. Sexually Transmitted Diseases. 1986, New York, USA. Churchill Livingstone; pp 305-307.
Fig. 1Colony (arrow) of T. vaginalis in soft agar 5 days after seeding at a concentration of 1,000 trichomonad per plate

Fig. 2Southern hybridization of 337 bp probe of T. vaginalis with EcoRI-digested trichomonad DNA. (A) Autoradiogram of Southern blot. Lane 1. KT8; 2, KT8C1; 3, KT8C2; 4, CDC85C; 5, IR78; 6, IR78C1; 7. IR78C2; 8, KT-Lee; 9, KT-LeeC2; 10, KT6; 11, NYH286; 12, KT-Kim; 13, T. foetus KV (B) Diagrammatic representation of banding patterns. Lanes 1-13 were identical to panel A. NYH286 strain (III group) had a peculiar 6.2 kb (⋆).

Citations
Citations to this article as recorded by

- Population structure and genetic diversity of Trichomonas vaginalis clinical isolates in Australia and Ghana
Daniel S. Squire, Alan J. Lymbery, Jennifer Walters, Frances Brigg, Andrea Paparini, R.C. Andrew Thompson
Infection, Genetics and Evolution.2020; 82: 104318. CrossRef - Genotypic Variation in Trichomonas vaginalis Detected in South African Pregnant Women
Rennisha Chetty, Nonkululeko Mabaso, Nathlee Abbai
Infectious Diseases in Obstetrics and Gynecology.2020; 2020: 1. CrossRef - Genetic Diversity of Trichomonas Vaginalis Clinical Isolates According to Restriction Fragment Length Polymorphism Analysis of the 60-kDa Proteinase Gene
Hilda Hernández, Jorge Fraga, Ricardo Marcet, Annia Alba, Mabel Figueredo, Yenisey Alfonso, Lázara Rojas, Jorge Sarracent
Acta Parasitologica.2019; 64(2): 300. CrossRef - Genetic diversity inTrichomonas vaginalis
John C Meade, Jane M Carlton
Sexually Transmitted Infections.2013; 89(6): 444. CrossRef - Genetic Characterization of Trichomonas vaginalis Isolates by Use of Multilocus Sequence Typing
Denise C. Cornelius, D. Ashley Robinson, Christina A. Muzny, Leandro A. Mena, David M. Aanensen, William B. Lushbaugh, John C. Meade
Journal of Clinical Microbiology.2012; 50(10): 3293. CrossRef - Double-stranded RNA viral infection of Trichomonas vaginalis and correlation with genetic polymorphism of isolates
Jorge Fraga, Lazara Rojas, Idalia Sariego, Ayme Fernández-Calienes
Experimental Parasitology.2011; 127(2): 593. CrossRef - Genetic variability between Trichomonas vaginalis isolates and correlation with clinical presentation
Lazara Rojas, Jorge Fraga, Idalia Sariego
Infection, Genetics and Evolution.2004; 4(1): 53. CrossRef