Abstract
In a previous study, we reported our discovery of Acanthamoeba contamination in domestic tap water; in that study, we determined that some Acanthamoeba strains harbor endosymbiotic bacteria, via our molecular characterization by mitochondrial DNA restriction fragment length polymorphism (Mt DNA RFLP). Five (29.4%) among 17 Acanthamoeba isolates contained endosymbionts in their cytoplasm, as demonstrated via orcein staining. In order to estimate their pathogenicity, we conducted a genetic characterization of the endosymbionts in Acanthamoeba isolated from domestic tap water via 16S rDNA sequencing. The endosymbionts of Acanthamoeba sp. KA/WP3 and KA/WP4 evidenced the highest level of similarity, at 97% of the recently published 16S rDNA sequence of the bacterium, Candidatus Amoebophilus asiaticus. The endosymbionts of Acanthamoeba sp. KA/WP8 and KA/WP12 shared a 97% sequence similarity with each other, and were also highly similar to Candidatus Odyssella thessalonicensis, a member of the α-proteobacteria. The endosymbiont of Acanthamoeba sp. KA/WP9 exhibits a high degree of similarity (85-95%) with genus Methylophilus, which is not yet known to harbor any endosymbionts. This is the first report, to the best of our knowledge, to show that Methylophilus spp. can live in the cytoplasm of Acanthamoeba.
-
Key words: Acanthamoeba, endosymbiont, Methylophilus, Candidatus Amoebophilus asiaticus, Candidatus Odyssella thessalonicensis
INTRODUCTION
Acanthamoeba spp. are ubiquitous free-living amoebae which function as predators that control microbial communities; these species have also been recognized as human pathogens that cause keratitis or granulomatous amebic encephalitis [
1]. In addition, a variety of other clinically-related bacteria, including
Mycobacterium avium,
Vibrio cholerae,
Listeria monocytogenes,
Legionella pneumophila, and
Burkholderia cepacia, are capable of surviving and multiplying for a limited time within
acanthamoebae [
2-
6]. It has been suggested that growth in an amebic intracellular environment might help these bacteria to adapt and survive within mammalian phagocytic cells [
7-
9]. Moreover, the endosymbiosis of bacteria in amoebic cysts has also been demonstrated to be relatively resistant to harmful environmental conditions, including exposure to antibiotic agents [
10,
11].
Several studies concerning the long-term symbiotic relationship between
Acanthamoeba and bacteria have been conducted since the first such study in the 1970's. These obligate bacterial endosymbionts have been reported in approximately 24% of all
Acanthamoeba isolates [
12]. However, the majority of obligate endosymbiotic bacteria could not be cultured via standard microbiological techniques. Recently, many researchers have attempted to characterize the diversity and phylogeny of these obligate intracellular microorganisms via molecular identification techniques. In some studies, comparative analyses of 16S rRNA sequences have successfully demonstrated the presence of several endosymbionts in
Acanthamoeba, including
Legionella [
13], Chlamydiales [
14], Rickettsiales, or Caedibacter-related bacteria [
15,
16].
In a previous study, we detected
Acanthamoeba contamination in domestic tap water, and evaluated molecular characteristics of these specimens using 18S rDNA sequencing and mitochondrial (Mt) DNA RFLP techniques [
17]. We determined that some
Acanthamoeba strains harbored endosymbiotic bacteria, via characterization by Mt DNA RFLP. In this study, we have confirmed the presence of endosymbiotic bacteria, using transmission electron microscopy. In order to estimate their pathogenicity, we conducted a genetic characterization of the endosymbionts in
Acanthamoeba isolates from domestic tap water using 16S rDNA sequencing.
MATERIALS AND METHODS
Isolation and maintenance of Acanthamoeba
Total 17 isolates from domestic tap water samples and reference strains were reported previously [
17,
18], and these strains were maintained as previously described [
19].
Orcein staining was utilized to visualize the presence of endosymbionts within the
Acanthamoeba isolates, as described by Beier et al. [
15]. Thin films of
Acanthamoeba were prepared on slides and allowed to dry in air. The slides were placed in 1% KMnO
4 for 5 min, washed with water, then rinsed for 30 sec in 3% oxalic acid and finally, washed off in distilled water. The films were covered with a few drops of the orcein-carmine stain solution. The slides were subsequently examined 1 hr after staining, under a light microscope (Olympus IX70, Tokyo, Japan) using oil immersion objectives.
Trophozoites were washed 3 times with cold PBS and then pre-fixed for 2 hr with 4% glutaraldehyde in 0.1 M cacodylate buffer. After rinsing with 0.1 M cacodylate buffer, the sediment was post-fixed for 3 hr with 1% osmium tetroxide, rinsed twice with 0.1 M maleate buffer at pH 5.2, dehydrated with ethyl alcohol, and treated for 30 min with propylene oxide. The pellet was immersed overnight in propylene oxide-resin (1 : 1) with continuous shaking, and then embedded in resin and incubated overnight at 60℃. Ultrathin sections cut on a Reichert-Jung ultramicrotome were stained with uranyl acetate and lead citrate. The sections were observed under a transmission electron microscope (Hitachi H-7000, Tokyo, Japan).
Sequence analysis of 16S rDNA of endosymbionts and 18S rDNA of Acanthamoeba
In order to amplify the 16S rDNA of bacterial endosymbionts, the Mt DNA of each
Acanthamoeba was utilized as a template. The nucleotide sequences of the primers were as follows: forward 5'-TCGACAACAGAGTTTGATCCTGGCTCAG-3', reverse 5'-ATCCAAGCTTAAGGAGGTGATCCAGCC-3'. The 16S rDNA amplification reactions were conducted separately in a thermal capillary cycler using premix of 50 µl scale (Bioneer, Seoul, Korea), including 1.25 U
Taq polymerase (Bioneer). The PCR reactions were conducted for an initial 3 min at 94℃ followed by 30 cycles of 94℃ for 1.5 min, 50℃ for 1 min, 72℃ for 4 min, and a final elongation step of 7 min at 72℃. Full-length amoeba 18S rDNA genes were amplified and sequenced as previously reported [
20]. In brief, the PCR reactions were conducted for an initial 3 min at 94℃ followed by 30 cycles of 94℃ for 1 min, 58℃ for 1 min, 72℃ for 2.5 min, and a final 10 min elongation step at 72℃. The amplified PCR products were ligated into the T/A cloning vector pGEM-T easy System I (Promega, Madison, Wisconsin, USA), with subsequent transformation in
E. coli by each vector. DNA sequencing was performed by an outside company (MACROGEN, Seoul, Korea).
Nucleotide sequences were compared to those of published strains via BLAST searches [
21]. Clustal X [
22] was used to conduct pairwise alignment and to calculate the percentage of sequence dissimilarity. The percent similarity corresponds to the number of identical sites between the 2 isolates, aligned to each other as in the master alignment. Phylogeny was determined using the ClustalW2 program (
http://www.ebi.ac.uk/Tools/clustalw2/index.html) with a low gap penalty, and phylogenetic trees were constructed with TreeView computer software.
RESULTS
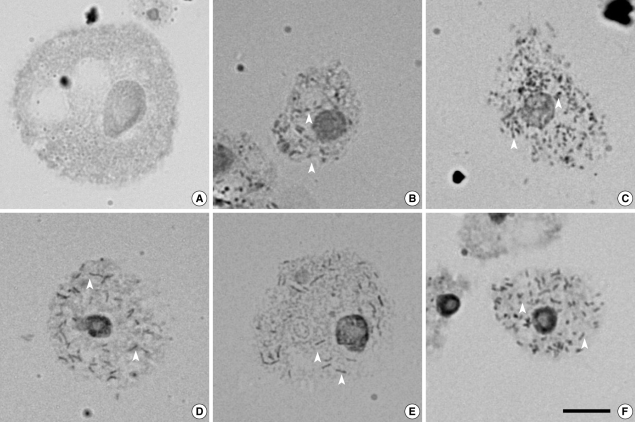
Endosymbionts in Acanthamoeba detected by orcein staining
Five isolates (29.4%) of
Acanthamoeba were found to harbor endosymbionts in their cytoplasm. The bacterial endosymbionts were readily visualized within
Acanthamoeba stained with orcein under phase-contrast light microscopy (
Fig. 1). Darkly stained rod-shaped bacterial endosymbionts were distributed randomly in the cytoplasm of trophozoites of
Acanthamoeba sp. KA/WP3, KA/WP4, KA/WP8, KA/WP9, and KA/WP12. In comparison, no bacteria were detected in
A. castellanii Castellani, which was employed as a reference strain.
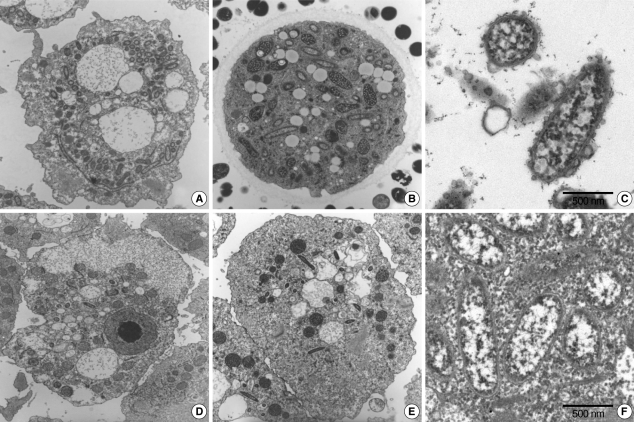
The morphological and ultrastructural characters of the endosymbionts were elucidated via transmission electron microscopy. We found that it was substantially easier to detect endosymbionts in the ameba than in
A. castellanii Castellani. The rodshaped endosymbionts detected in KA/WP3 and KA/WP4 were estimated to be approximately 1.72 (1.70-1.75) × 0.27 (0.20-0.30) µm in size (
Fig. 2A-C), and the endosymbionts (1.55 × 0.25 µm) detected in KA/WP8 and KA/WP12 were smaller than those observed in KA/WP3 and KA/WP4 (
Fig. 2D-F). The endosymbionts of KA/WP3 and KA/WP4 were quite similar to each other in terms of morphological characteristics, and they could be readily detected in the cytoplasm of
Acanthamoeba, because they existed at numbers in excess of 500 per
Acanthamoeba. Additionally, we noted that these endosymbionts were bound by double membranes and their cell surfaces were studded with numerous host cell ribosomes (
Fig. 2C).
We also noted some different morphological characteristics in the endosymbionts found in KA/WP8 and KA/WP12; the endosymbionts in these strains were bound by double membranes, but also exhibited apparent vesicle-like structures on the bacterial outer membrane; a translucent zone surrounding the endosymbiont was also observed (
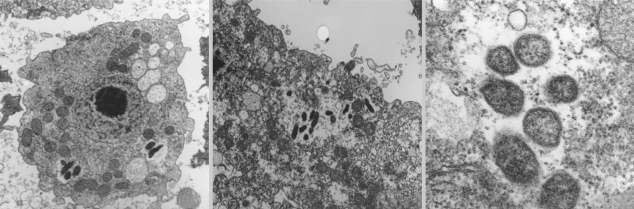
Fig. 2F). Additionally, the endosymbionts of KA/WP3, KA/WP4, KA/WP8, and KA/WP12 were found to be distributed randomly throughout the cytoplasm of the trophozoites and cyst forms, but the endosymbionts of KA/WP9 were distributed with group (2-16 endosymbionts per group) in the cytoplasm (
Fig. 3). However, these endosymbionts are not surrounded by any membranes or vesicles. The endosymbionts of KA/WP9 also evidenced a very different shape than the other endosymbionts in this study; their bodies were curved rod-shaped and quite irregular (asymmetry rod shape).
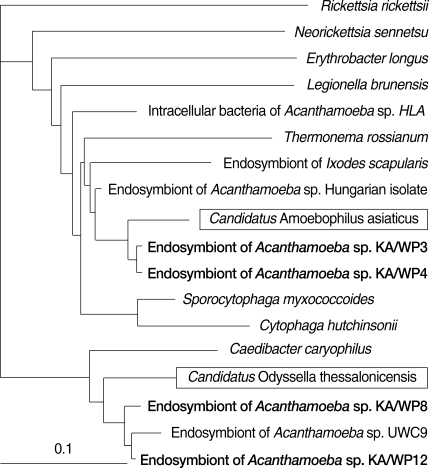
Comparative analysis with 16S rRNA gene sequences demonstrated that the endosymbionts of KA/WP3 and KA/WP4 evidenced 99% sequence similarity with each other. The endosymbionts of KA/WP3 and KA/WP4 showed the highest similarity, at 97% of the recently published 16S rDNA sequence of the bacteria,
Candidatus Amoebophilus asiaticus [
23]. Additionally, these symbionts evidenced a high degree of similarity with the endosymbionts of the tick,
Ixodes scapularis (89% and 90%, respectively). The phylogenetic analysis consistently demonstrated that the endosymbiont of KA/WP3 and KA/WP4,
Candidatus Amoebophilus asiaticus, and the endosymbiont of
I. scapularis form a monophyletic lineage within the CFB phylum (
Fig. 4). However, the endosymbiont of KA/WP3 and KA/ WP4 demonstrated only moderate similarity (less than 80%) with the other endosymbionts detected in KA/WP8, KAWP9, and KAWP12. Comparative analysis with 16S rRNA gene sequences showed that the endosymbionts of KA/WP8 and KA/WP12 shared 97% sequence similarity with each other. These endosymbionts evidenced a high degree of similarity to
Candidatus Odyssella thessalonicensis, a member of the α-proteobacteria. They share an evolutionary line of descent with
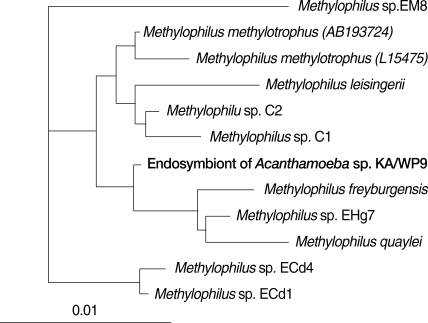
Caedibacter caryophilus. The endosymbiont of KA/WP9 is highly similar (85-95%) to genus
Methylophilus, which has not previously been determined to harbor any endosymbionts (
Fig. 5). This is the first report to show that
Methylophilus spp. can live in the cytoplasm of
Acanthamoeba.
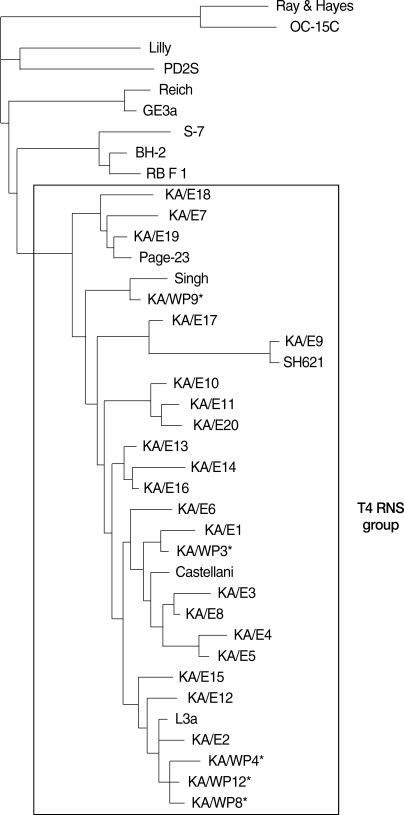
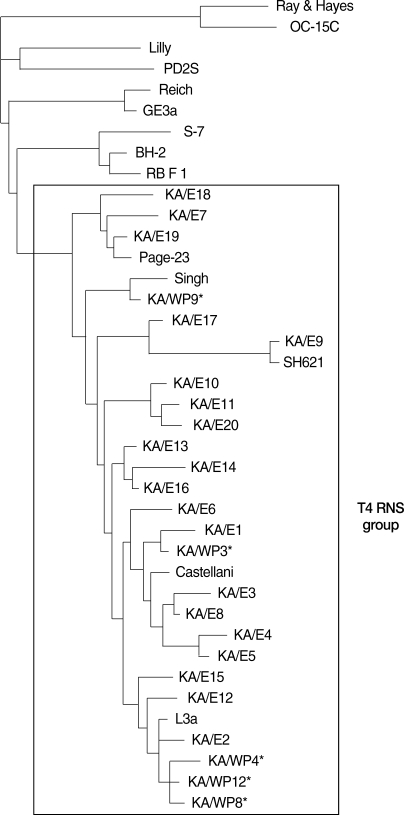
The dendrogram constructed on the basis of the 18S rDNA sequence analyses is shown in
Fig. 6. Comparative sequence analysis verified the morphology-based identification of this isolate as
Acanthamoeba sp. KA/WP3 evidenced the highest level of sequence similarity (98%) with
A. castellanii Castellani. In addition,
Acanthamoeba KA/WP3 evidenced a high 18S rDNA sequence similarity (99%) with the clinical isolates of KA/E1 (isolated from Korean patients with keratitis).
Acanthamoeba KA/WP4, KA/WP8, and KA/WP12 exhibited the highest level of sequence similarity (98%) with
A. lugdunensis L3a. These amebas also exhibited very high 18S rDNA sequence similarity (97%) with the clinical isolates KA/E2, KA/E12, KA/E15.
Acanthamoeba KA/WP9 is closely related to
A. rhysodes Singh, which was isolated from soil. This amoeba also has not previously been recognized as a host for any endosymbionts. Using the 95% similarity value for the definition of the
Acanthameoba 18S rDNA sequence types [
24], all of the host
Acanthamoeba in this study could be assigned to genotype T4, which comprises the majority of clinical and environmental
Acanthamoeba isolates.
DISCUSSION
Bacterial endosymbionts are commonly observed in protozoans, particularly for
Acanthamoeba; approximately 24% of
Acanthamoeba isolates from both environmental and medical samples were determined to harbor endosymbionts [
12,
25]. In our study, 29.4% of
Acanthamoeba isolated from domestic tap water harbored endosymbiont bacteria in their cytoplasm.
A number of endosymbionts of both KA/WP3 and KA/WP4 were detected in the cytoplasm of
Acanthamoeba, and they were identified in this study as
Candidatus Amoebophilus asiaticus, via 16S rDNA sequencing analysis. This bacteria was initially described by Horn et al. [
23], who explained that "Amoebophilus" taxonomically designates the obligate intracellular lifestyle of the bacteria in free-living amoebae, and the term "asiaticus" refers to continental Asia, where the host was originally isolate. Two independent laboratories previously attempted to culture the bacteria in a number of cell-free media, but they failed [
23]. These findings suggested that eukaryotic host cells are essential for the growth of these bacteria, similarly to mitochondria. We also speculate that the endosymbiont did not affect, or only minimally affected the biological functions of the ameba, because a number of bacteria (especially endosymbionts of KA/WP3) occupied the ameba cytoplasm although there were no pathological findings (
Fig. 2A). These endosymbionts were surrounded by a double membrane decorated with numerous ribosomes of the host
Acanthamoeba. These morphological characteristics are consistent with those reported for the endosymbionts of
Acanthamoeba isolated previously from contact lens storage cases in Korea and other countries [
26-
28].
We also identified other obligate intracellular bacteria detected in KA/WP8 and KA/WP12 as Candidatus Odyssella thessalonicensis. This species name was suggested by Birtle et al. [
28], and referenced Odysseus and Thessalonika (the Greek city). These bacteria cannot be cultivated on cell-free medium, and can only be co-cultivated with a number of
Acanthamoeba species at temperatures at or below 37℃. At temperatures above 30℃, the organism multiplies and rapidly destroys its host [
28]. We noted an electron-translucent area surrounding the bacteria, which was attributed to the presence of bacterial capsular material. Similar endosymbionts were also reported [
28,
29].
In this study, we identified an endosymbiont from KA/WP9, the morphological and molecular characteristics of which were quite similar to those of
Methylophilus spp.
Methylophilus was suggested as a new genus by Jankins et al. in 1987; they described the characterization of the bacteria as follows [
30]. When grown on methanol-mineral salts agar or in methanol mineral salts liquid medium, straight, or slightly curved rods usually 0.3 to 0.6 pm in diameter and 0.8 to 1.5 pm in length occur either singly or in pairs; they are gram-negative, but the stain is frequently not taken up well. They are motile (by polar flagella) or non-motile. Endospores are absent; no cellular inclusions; no sheath or prosthecae detected; no capsules formed, but slime may be produced by some strains [
30]. The bacteria occur singly or in pairs and can be isolated from sludge, mud, and river and pond water [
31]. Because their niche is shared with
Acanthamoeba, they can be fed by
Acanthamoeba. However, until now,
Methylophilus has not previously been recognized as an endosymbiont occupying
Acanthamoeba.
We identified endosymbionts (
Methylophilus spp.) from KA/WP9 using electron microscopy after 6 months from axenic culture. We could check for endosymbionts from KA/WP9 until 1 year after identification. However, the KA/WP9 culture medium became contaminated by unknown bacteria, and thus we replaced the culture medium with one containing antibiotics. Afterward, we were unable to detect any endosymbionts in KA/WP9. We are still uncertain as to whether the contaminating bacteria were endosymbionts or not. The endosymbionts of KA/WP9 was not surrounded by host phagocytic membranes (
Fig. 3F), and only a few bacteria were detected in the cytoplasm of KA/WP9 (
Fig. 3E). We believe that this stage may be an initial step in the symbiosis between the amoeba and the bacteria.
Many medically important bacteria are capable of utilizing
Acanthamoeba as shelter; in particular, many studies have focused on
Legionella and
Mycobacterium in this regard [
32,
33]. We don't know whether or not the endosymbiont of KA/WP9, Methylophilus, is virulent, because we failed in our attempts to cultivate these bacteria with Methylophilus-specific medium. With the exception of the presence of endosymbionts,
Acanthamoeba sp. KA/WP3 and KA/WP4 with
Acanthamoeba sp. KA/E1 (keratitis isolates), as well as KA/WP8 and KA/WP12 with
A. lugdunensis L3a and
Acanthamoeba sp. KA/E2 were phylogenetically closely related, according to the results of Mt DNA RFLP and 18S rDNA sequence analysis [
17]. They are most frequently isolated from contact lens storage cases and keratitis patients in Korea. This isogenetic pair can be utilized to evaluate the relationship of the bacteria with their host,
Acanthamoeba. Further studies should focus on the functions of these endosymbionts, which enhance amoebic cytopathogenicity.
ACKNOWLEDGEMENT
This work was supported for 2 years by Pusan National University Research Grant.
References
- 1. Schuster FL, Visvesvara GS. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int J Parasitol 2004;34:1001-1027.
- 2. Miltner EC, Bermudez LE. Mycobacterium avium grown in Acanthamoeba castellanii is protected from the effects of antimicrobials. Antimicrob Agents Chemother 2000;44:1990-1994.
- 3. Abd H, Saeed A, Weintraub A, Nair GB, Sandström G. Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol Ecol 2007;60:33-39.
- 4. Ly TM, Müller HE. Ingested Listeria monocytogenes survive and multiply in protozoa. J Med Microbiol 1990;33:51-54.
- 5. Neumeister B. Intracellular multiplication of Legionella species and the influence of amoebae on their intracellular growth in human monocytes: mono mac 6 cells and Acanthamoeba castellanii as suitable in vitro models. Methods Mol Biol 2004;268:141-151.
- 6. Landers P, Kerr KG, Rowbotham TJ, Tipper JL, Keig PM, Ingham E, Denton M. Survival and growth of Burkholderia cepacia within the free-living amoeba Acanthamoeba polyphaga. Eur J Clin Microbiol Infect Dis 2000;19:121-123.
- 7. Tenant R, Bermudez LE. Mycobacterium avium genes upregulated upon infection of Acanthamoeba castellanii demonstrate a common response to the intracellular environment. Curr Microbiol 2006;52:128-133.
- 8. Abd H, Saeed A, Weintraub A, Sandström G. Vibrio cholerae O139 requires neither capsule nor LPS O side chain to grow inside Acanthamoeba castellanii. J Med Microbiol 2009;58:125-131.
- 9. Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun 1997;65:3759-3767.
- 10. Barker J, Scaife H, Brown MR. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob Agents Chemother 1995;39:2684-2688.
- 11. Zhou X, Elmose J, Call DR. Interactions between the environmental pathogen Listeria monocytogenes and a free-living protozoan (Acanthamoeba castellanii). Environ Microbiol 2007;9:913-922.
- 12. Fritsche TR, Gautom RK, Seyedirashti S, Bergeron DL, Lindquist TD. Occurrence of bacterial endosymbionts in Acanthamoeba spp. isolated from corneal and environmental specimens and contact lenses. J Clin Microbiol 1993;31:1122-1126.
- 13. Birtles RJ, Rowbotham TJ, Raoult D, Harrison TG. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology 1996;142:3525-3530.
- 14. Corsaro D, Feroldi V, Saucedo G, Ribas F, Loret JF, Greub G. Novel Chlamydiales strains isolated from a water treatment plant. Environ Microbiol 2009;11:188-200.
- 15. Beier CL, Horn M, Michel R, Schweikert M, Gortz HD, Wagner M. The genus Caedibacter comprises endosymbionts of Paramecium spp. related to the Rickettsiales (Alphaproteobacteria) and to Francisella tularensis (Gammaproteobacteria). Appl Environ Microbiol 2002;68:6043-6050.
- 16. Horn M, Fritsche TR, Gautom RK, Schleifer KH, Wagner M. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus. Environ Microbiol 1999;1:357-367.
- 17. Jeong HJ, Lee SJ, Kim JH, Xuan YH, Lee KH, Park SK, Choi SH, Chung DI, Kong HH, Ock MS, Yu HS. Acanthamoeba: keratopathogenicity of isolates from domestic tap water in Korea. Exp Parasitol 2007;117:357-367.
- 18. Jeong HJ, Yu HS. The role of domestic tap water in Acanthamoeba contamination in contact lens storage cases in Korea. Korean J Parasitol 2005;43:47-50.
- 19. Yu HS, Choi KH, Kim HK, Kong HH, Chung DI. Genetic analyses of Acanthamoeba isolates from contact lens storage cases of students in Seoul, Korea. Korean J Parasitol 2001;39:161-170.
- 20. Chung DI, Yu HS, Hwang MY, Kim TH, Kim TO, Yun HC, Kong HH. Subgenus classification of Acanthamoeba by riboprinting. Korean J Parasitol 1998;36:69-80.
- 21. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215:403-410.
- 22. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;25:4876-4882.
- 23. Horn M, Harzenetter MD, Linner T, Schmid EN, Müller KD, Michel R, Wagner M. Members of the Cytophaga-Flavobacterium-Bacteroides phylum as intracellular bacteria of acanthamoebae: proposal of 'Candidatus Amoebophilus asiaticus'. Environ Microbiol 2001;3:440-449.
- 24. Stothard DR, Schroeder-Diedrich JM, Awwad MH, Gast RJ, Ledee DR, Rodriguez-Zaragoza S, Dean CL, Fuerst PA, Byers TJ. The evolutionary history of the genus Acanthamoeba and the identification of eight new 18S rRNA gene sequence types. J Eukaryot Microbiol 1998;45:45-54.
- 25. Horn M, Wagner M. Bacterial endosymbionts of free-living amoebae. J Eukaryot Microbiol 2004;51:509-514.
- 26. Chung DI, Kong HH, Kim TH, Hwang MY, Yu HS, Yun HC, Seol SY. Bacterial endosymbiosis within the cytoplasm of Acanthamoeba lugdunensis isolated from a contact lens storage case. Korean J Parasitol 1997;35:127-133.
- 27. Yu HS, Jeong HJ, Hong YC, Seol SY, Chung DI, Kong HH. Natural occurrence of Mycobacterium as an endosymbiont of Acanthamoeba isolated from a contact lens storage case. Korean J Parasitol 2007;45:11-18.
- 28. Birtles RJ, Rowbotham TJ, Michel R, Pitcher DG, Lascola B, Alexiou-Daniel S, Raoult D. 'Candidatus Odyssella thessalonicensis' gen. nov., sp. nov., an obligate intracellular parasite of Acanthamoeba species. Int J Syst Evol Microbiol 2000;50:63-72.
- 29. Hall J, Voelz H. Bacterial endosymbionts of Acanthamoeba sp. J Parasitol 1985;71:89-95.
- 30. Jenkins O, Byrom D, Jones D. Methylophilus: a new genus of methanol-utilizing bacteria. Int J Syst Bacteriol 1987;37:446-448.
- 31. Bergey DH, Holt JG. Bergey's manual of determinative bacteriology. 1994, 9th ed. Baltimore, USA. Williams & Wilkins.
- 32. Steinert M, Birkness K, White E, Fields B, Quinn F. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl Environ Microbiol 1998;64:2256-2261.
- 33. Hay J, Seal DV, Billcliffe B, Freer JH. Non-culturable Legionella pneumophila associated with Acanthamoeba castellanii: detection of the bacterium using DNA amplification and hybridization. J Appl Bacteriol 1995;78:61-65.
Fig. 1Endosymbionts were detected in 5 (29%) of 17 Acanthamoeba isolates from domestic tap water using an orcein staining method. Arrow heads indicate endosymbionts. (A) Acanthamoeba castellanii Castellani. (B) Acanthamoeba sp. KA/WP3. (C) Acanthamoeba sp. KA/WP4. (D) Acanthamoeba sp. KA/WP8. (E) Acanthamoeba sp. KA/WP12. (F) Acanthamoeba sp. KA/WP9. Bar = 10 µm.

Fig. 2The endosymbiont structure was detected in Acanthamoeba sp. cytoplasm by electromicroscopy analysis. (A & C) Acanthamoeba sp. KA/WP3. (B) Acanthamoeba sp. KA/WP4. (D & F) Acanthamoeba sp. KA/WP8. (E) Acanthamoeba sp. KA/WP12.

Fig. 3New endosymbiont detected in Acanthamoeba sp. KA/WP9 isolated from domestic tap water. They are not surrounded by any membrane and most of the endosymbionts existed as a troop, not alone, in the cytoplasm of Acanthamoeba.
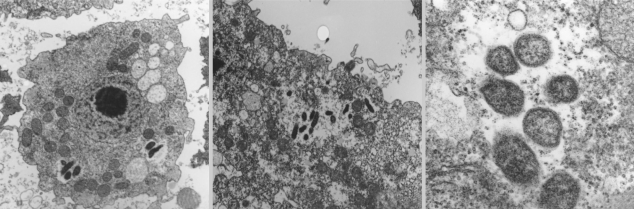
Fig. 416S rRNA-based neighbor-joining tree reflecting the affiliation of endosymbionts of Acanthamoeba sp. KA/WP3, KA/WP4, KA/WP8, and KA/WP12 with selected endosymbionts bacteria.

Fig. 516S rRNA-based neighbor-joining tree of endosymbiont of Acanthamoeba sp. KA/WP9 with members of Methylophilus.
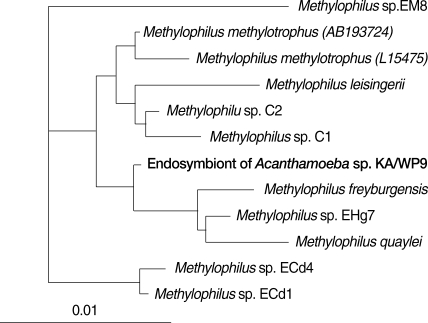
Fig. 6The reference with their corresponding accession numbers for Acanthamoeba 18S rDNA sequences used in this study are A. astronyxis Ray & Hayes, AF019064; A. tubiash OC-15C, AF019065; A. lenticulata PD2S, U94741; A. culbertsoni Lilly A-1, AF019067; A. healyi OC-3A, AF019070; A. palestinensis Reich, U07411; A. pustulosa GE3a, AF019050; A. griffini S-7, U07412; A. hatchetti BH-2, AF019068; A. stevensoni RB-F-1, AF019069; A. polyphaga Page, AF019061; A. triangularis SH621, AF316547; A. rhysodes Singh, AF351644; A. mauritaniensis 1652, AY351647; A. divionensis AA2, AY351646; A. paradivionensis AA1, AY351645; A. castellanii castellani, U07413; A. lugdunensis L3a, AF005995; A. castellanii Neff, AF690457; A. castellanii Ma, U07414 strain KA/E2, AF005998; KA/E6, AF349044; KA/E9, AF316546; KA/E12, AF316545; KA/E15, AY14-8961; KA/E16, AY148962.; KA/E17, AY148963; KA/E18, AY148964.